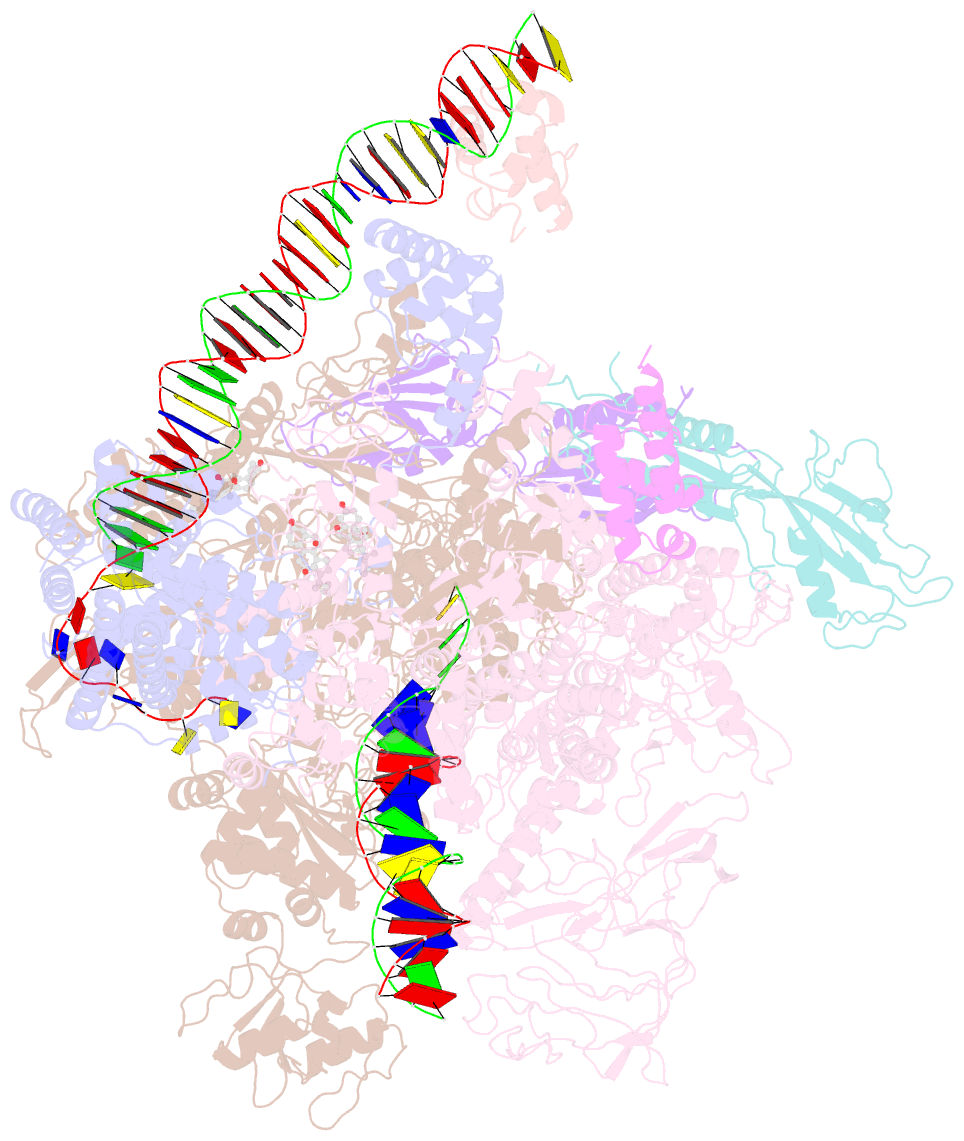
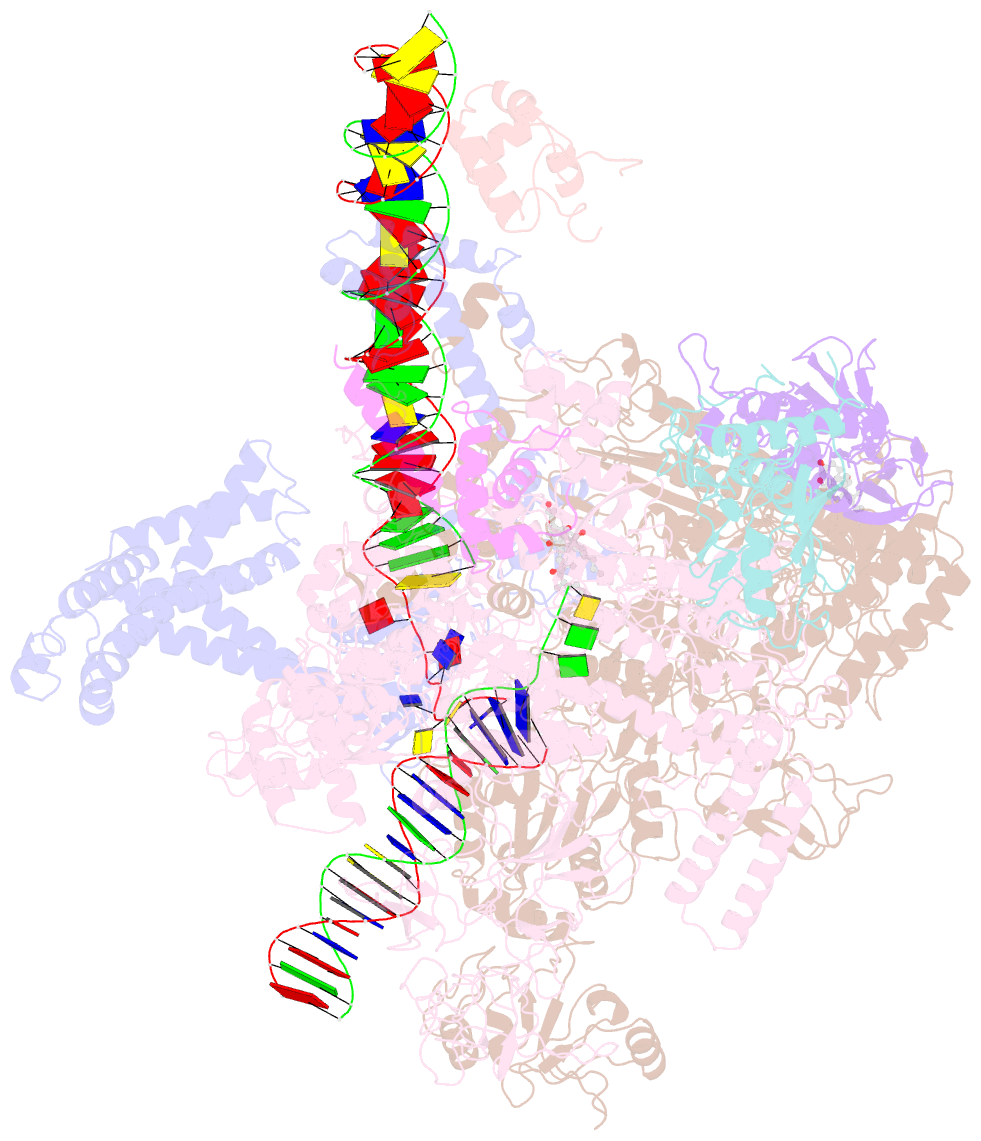
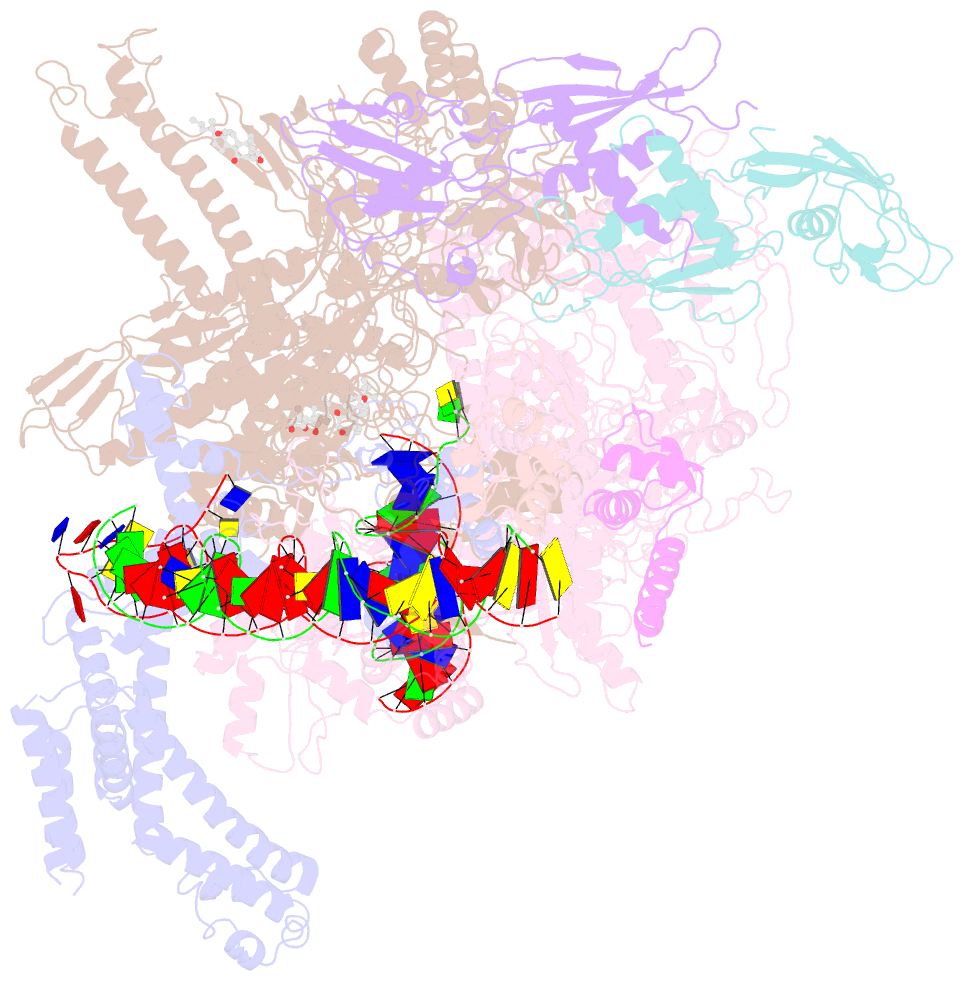
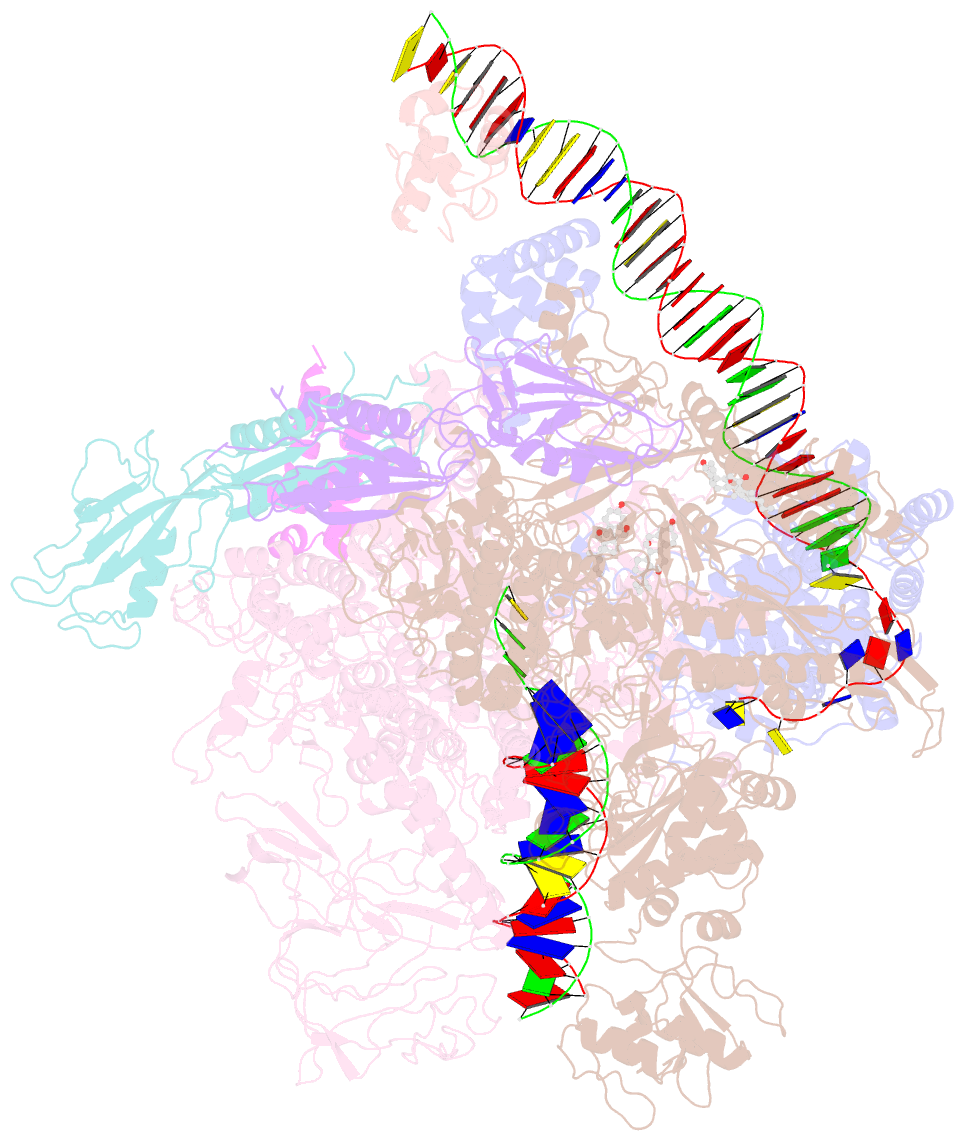
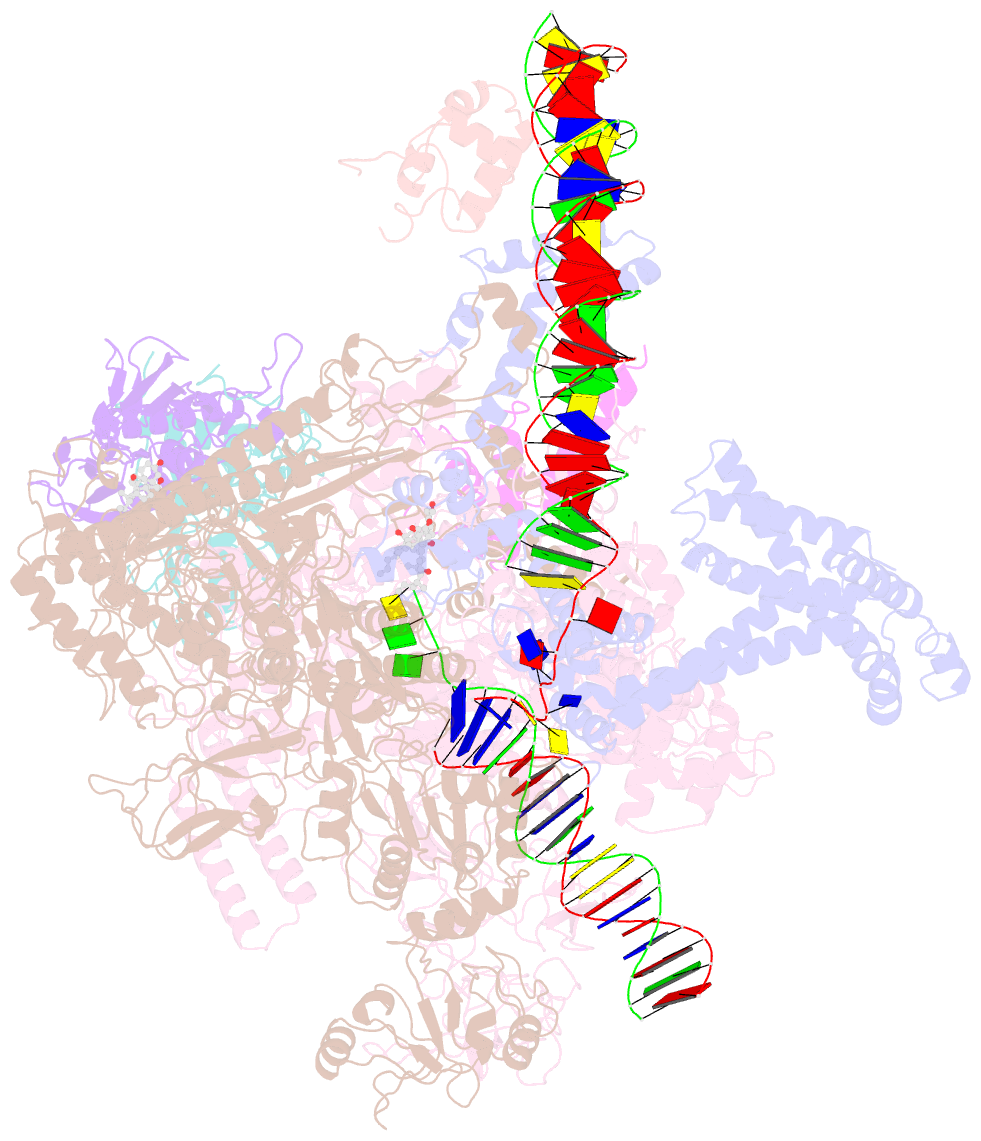
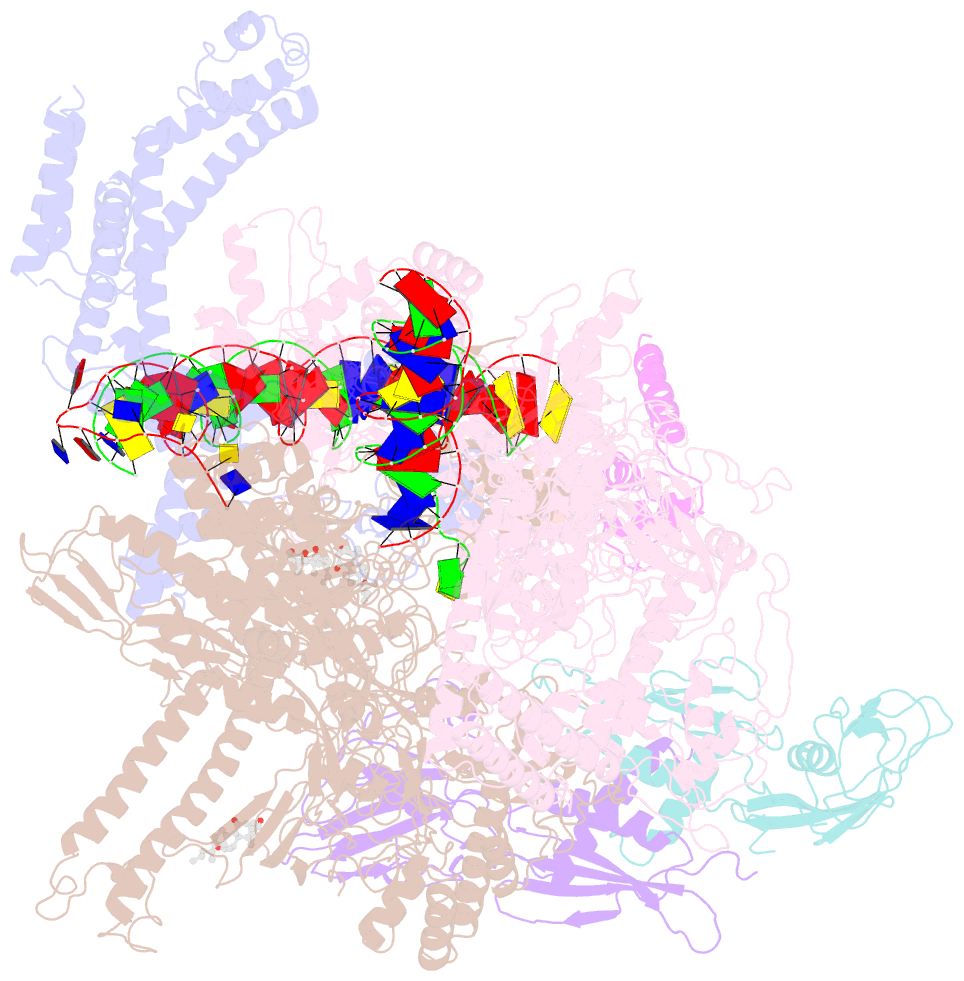
Summary information and primary citation
- PDB-id
- 6oul; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.4 Å)
- Summary
- cryo-EM structure of escherichia coli rnap polymerase bound to rpstp2 promoter DNA
- Reference
- Chen J, Gopalkrishnan S, Chiu C, Chen AY, Campbell EA, Gourse RL, Ross W, Darst SA (2019): "E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation." Elife, 8. doi: 10.7554/eLife.49375.
- Abstract
- TraR and its homolog DksA are bacterial proteins that regulate transcription initiation by binding directly to RNA polymerase (RNAP) rather than to promoter DNA. Effects of TraR mimic the combined effects of DksA and its cofactor ppGpp, but the structural basis for regulation by these factors remains unclear. Here, we use cryo-electron microscopy to determine structures of Escherichia coli RNAP, with or without TraR, and of an RNAP-promoter complex. TraR binding induced RNAP conformational changes not seen in previous crystallographic analyses, and a quantitative analysis revealed TraR-induced changes in RNAP conformational heterogeneity. These changes involve mobile regions of RNAP affecting promoter DNA interactions, including the βlobe, the clamp, the bridge helix, and several lineage-specific insertions. Using mutational approaches, we show that these structural changes, as well as effects on σ70 region 1.1, are critical for transcription activation or inhibition, depending on the kinetic features of regulated promoters.