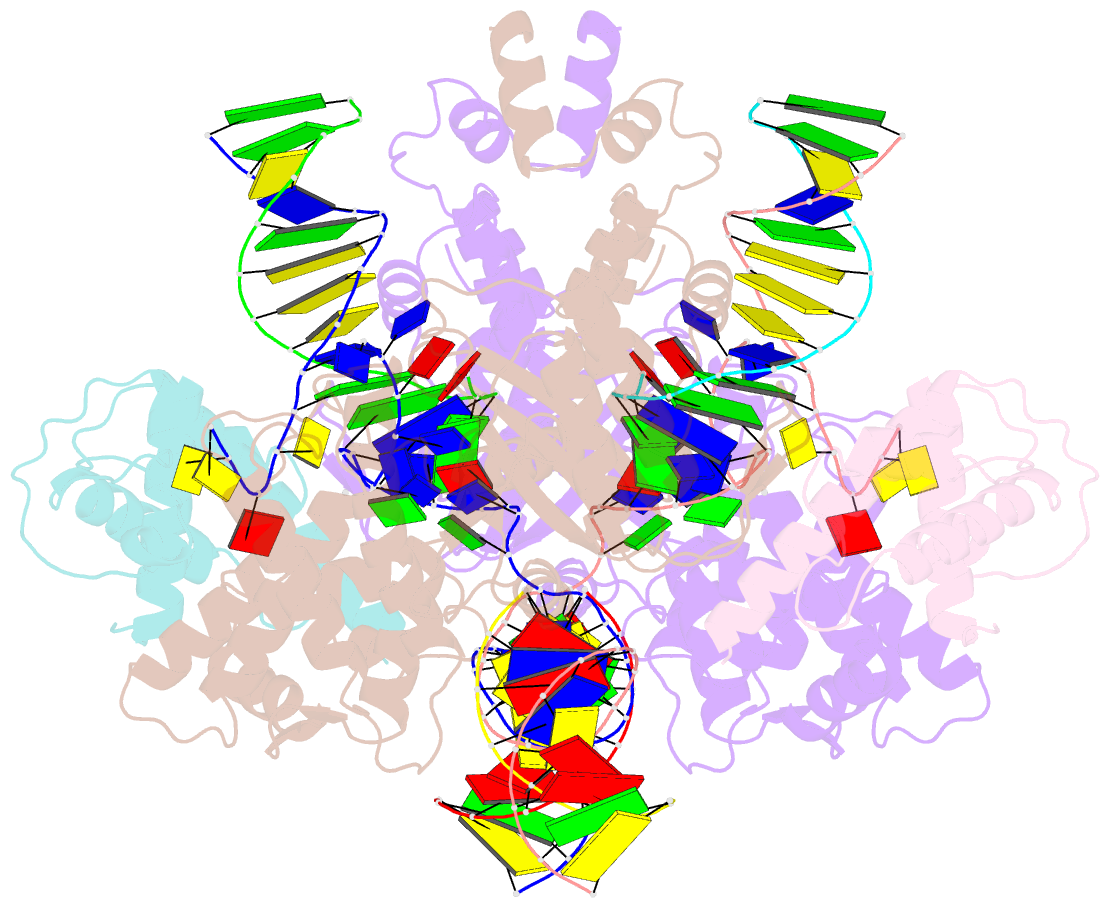
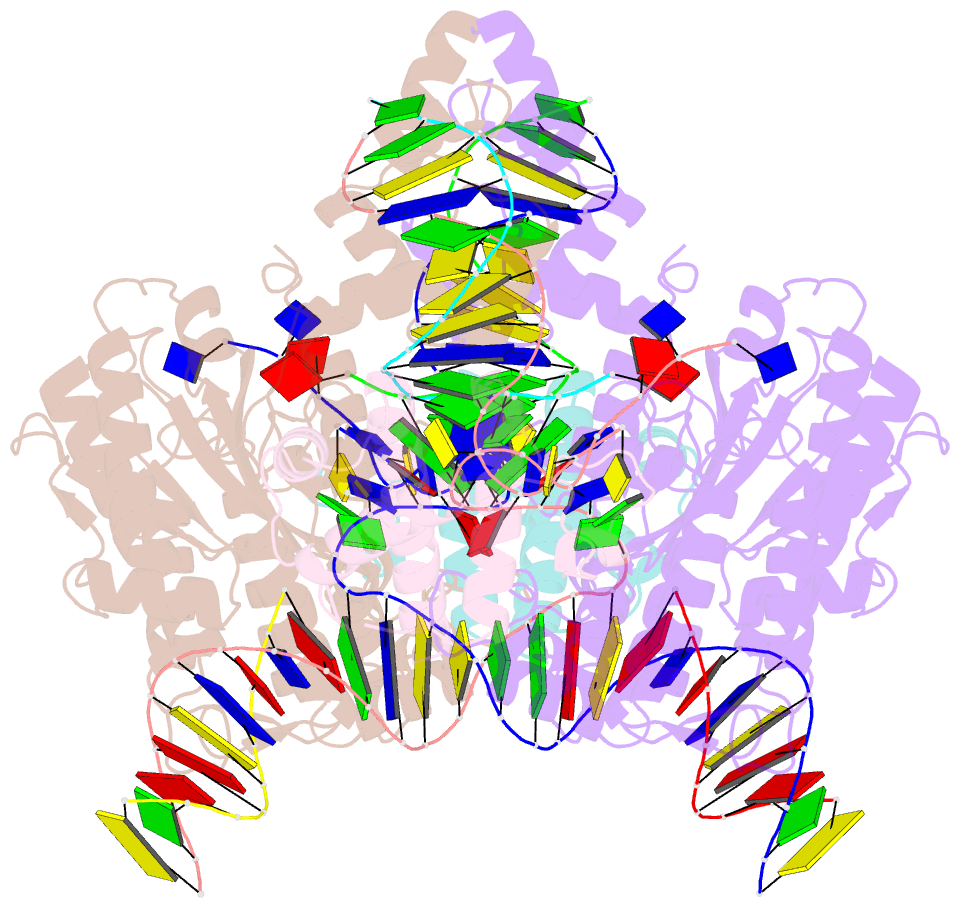
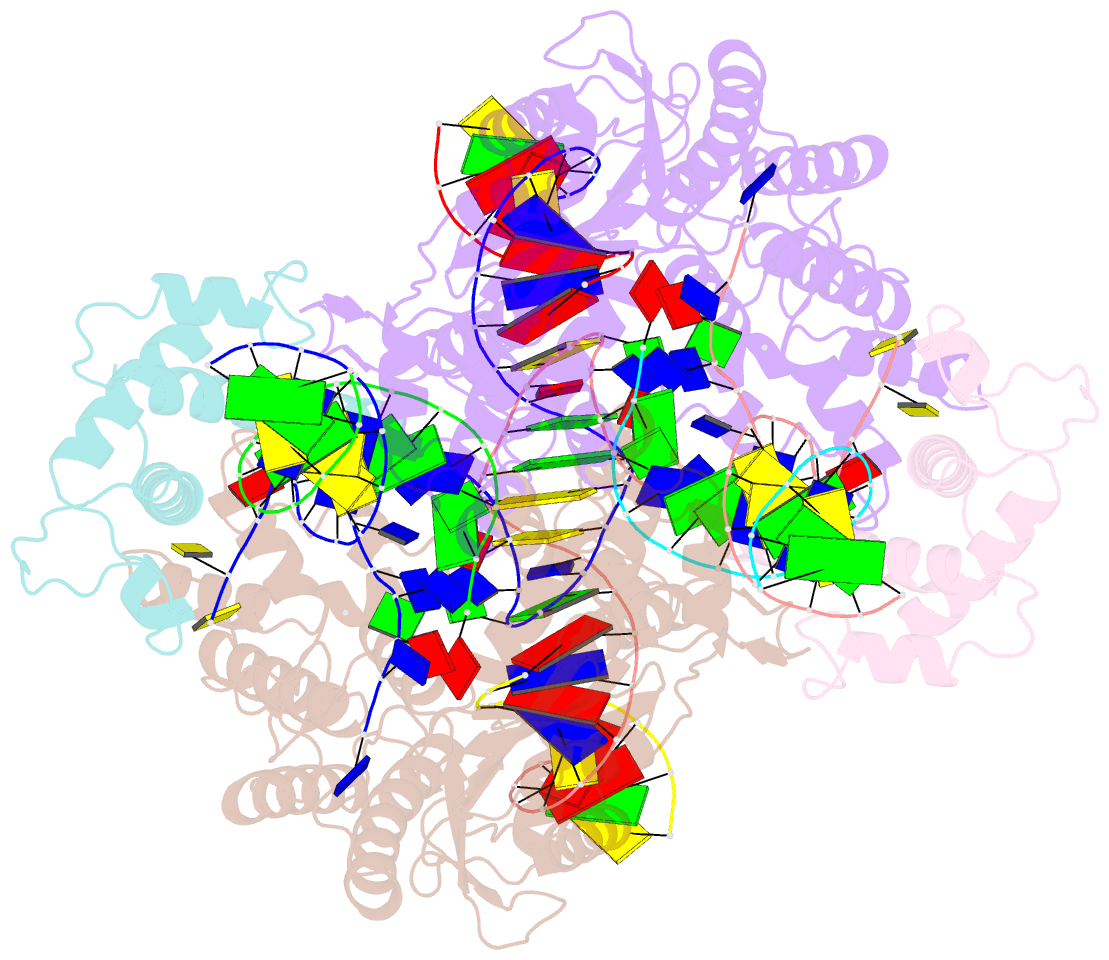
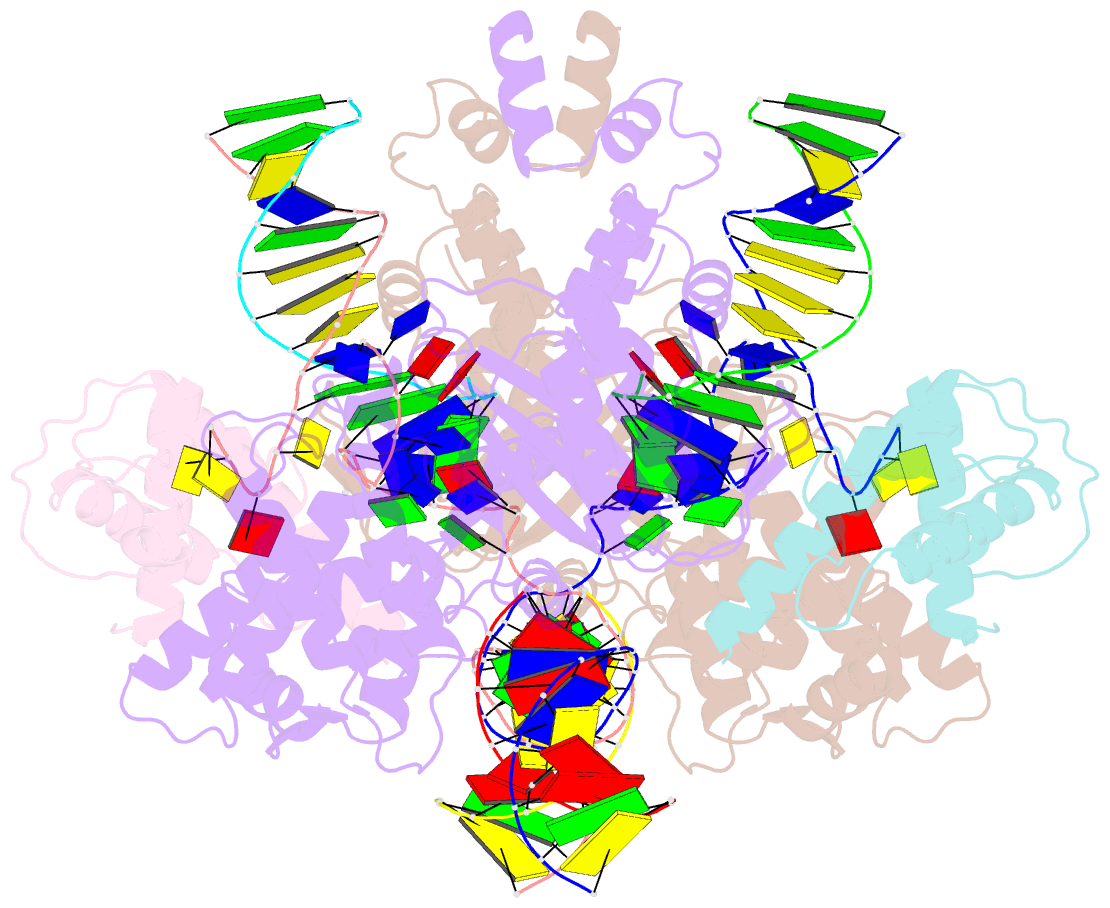
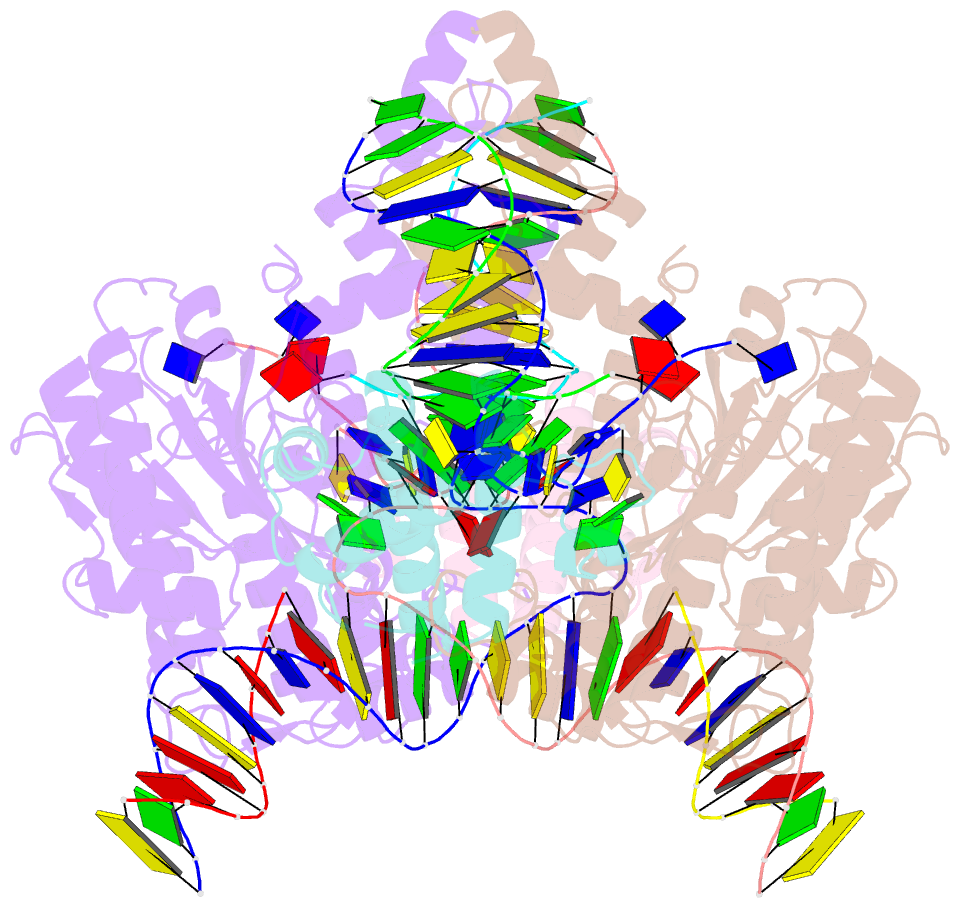
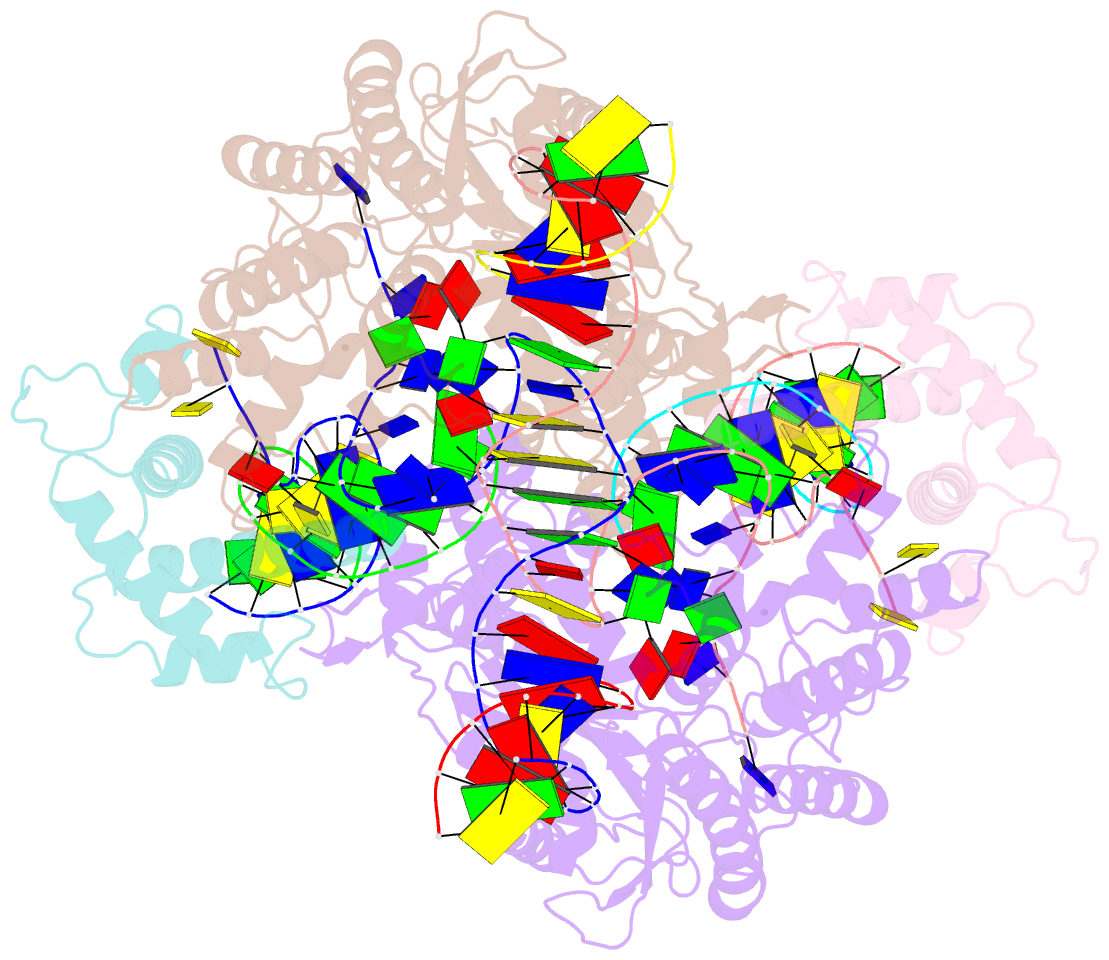
Summary information and primary citation
- PDB-id
- 6p5a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- cryo-EM (3.6 Å)
- Summary
- Drosophila p element transposase strand transfer complex
- Reference
- Ghanim GE, Kellogg EH, Nogales E, Rio DC (2019): "Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts." Nat.Struct.Mol.Biol., 26, 1013-1022. doi: 10.1038/s41594-019-0319-6.
- Abstract
- P element transposase catalyzes the mobility of P element DNA transposons within the Drosophila genome. P element transposase exhibits several unique properties, including the requirement for a guanosine triphosphate cofactor and the generation of long staggered DNA breaks during transposition. To gain insights into these features, we determined the atomic structure of the Drosophila P element transposase strand transfer complex using cryo-EM. The structure of this post-transposition nucleoprotein complex reveals that the terminal single-stranded transposon DNA adopts unusual A-form and distorted B-form helical geometries that are stabilized by extensive protein-DNA interactions. Additionally, we infer that the bound guanosine triphosphate cofactor interacts with the terminal base of the transposon DNA, apparently to position the P element DNA for catalysis. Our structure provides the first view of the P element transposase superfamily, offers new insights into P element transposition and implies a transposition pathway fundamentally distinct from other cut-and-paste DNA transposases.