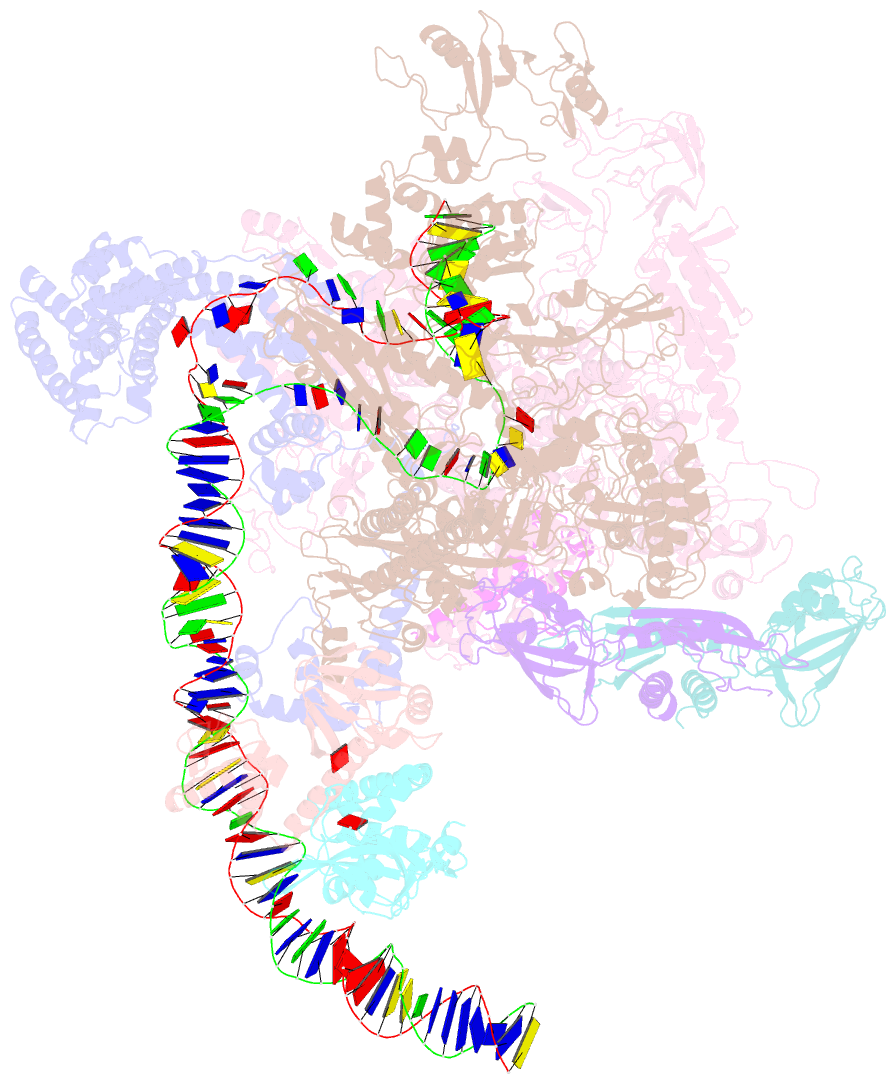
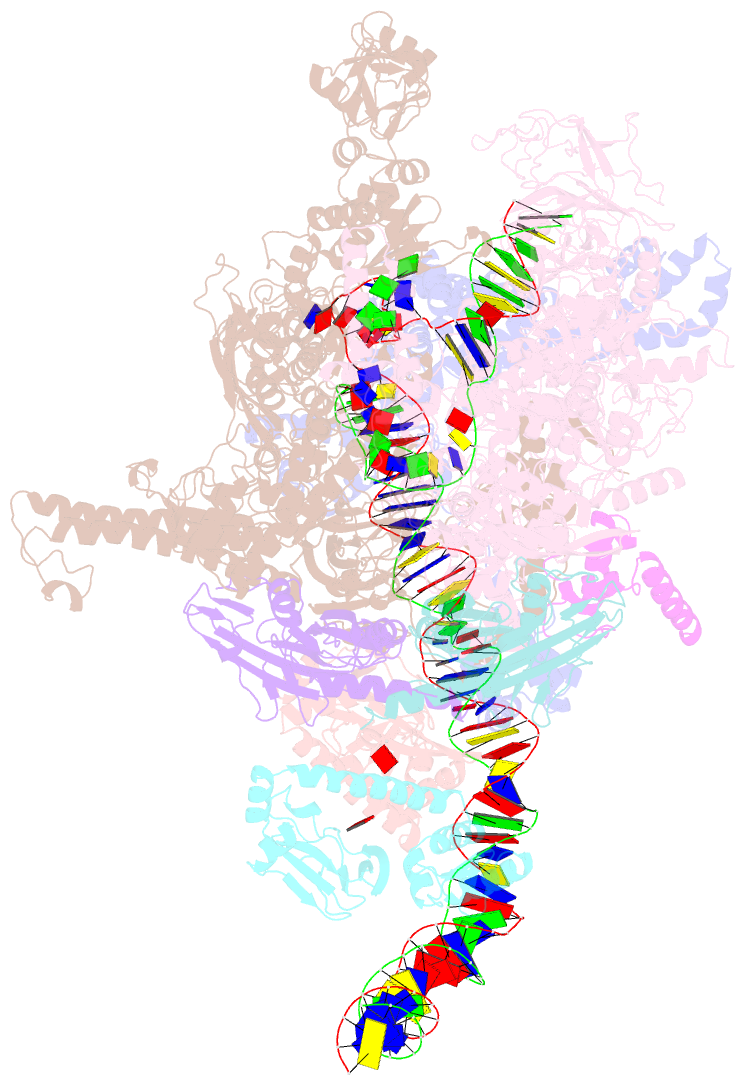
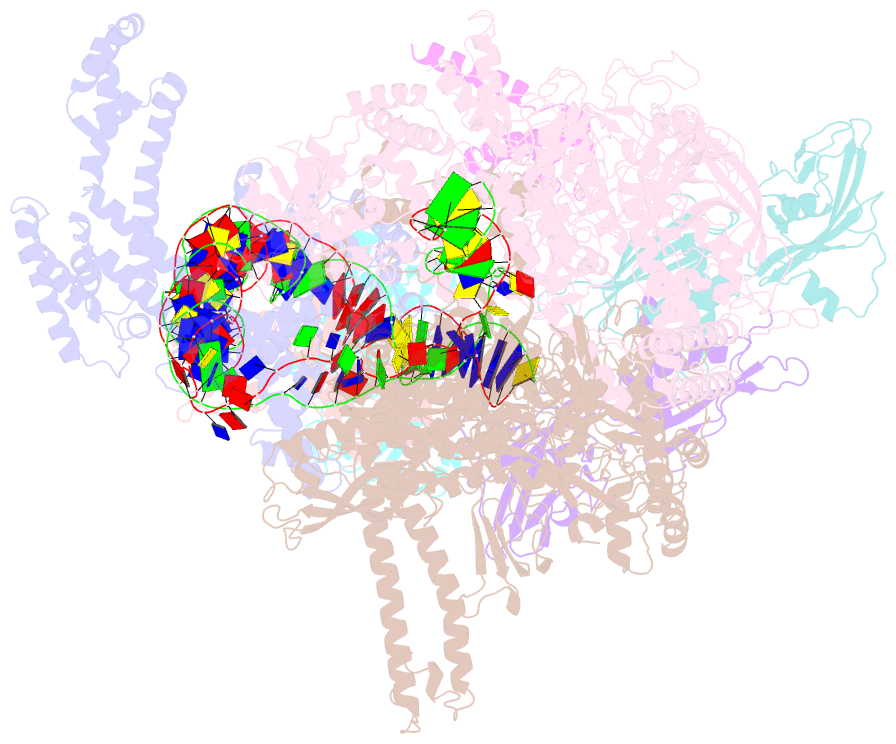
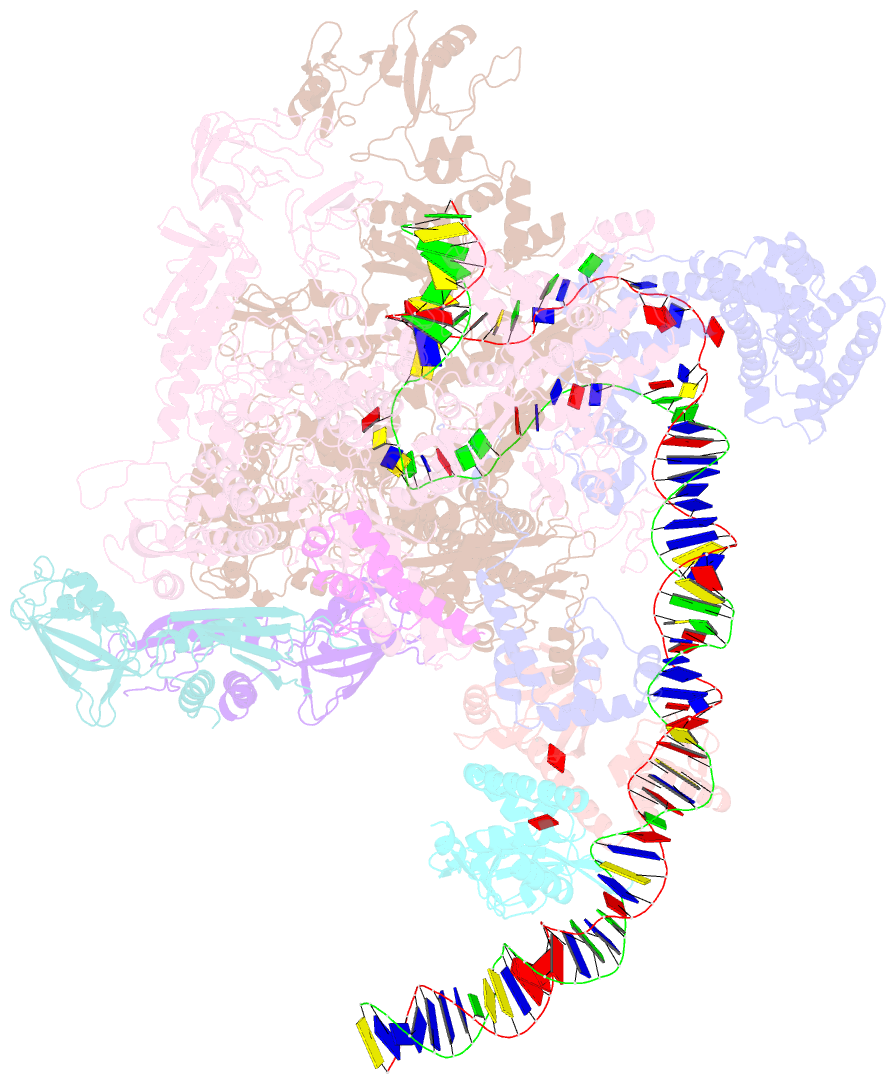
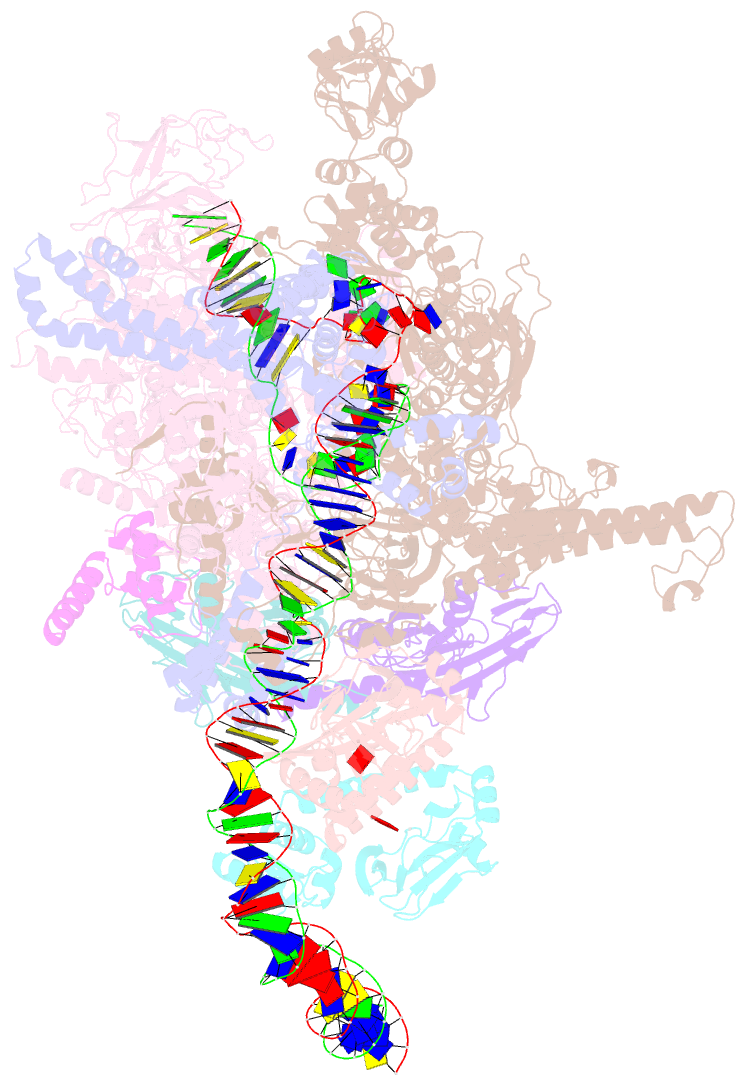
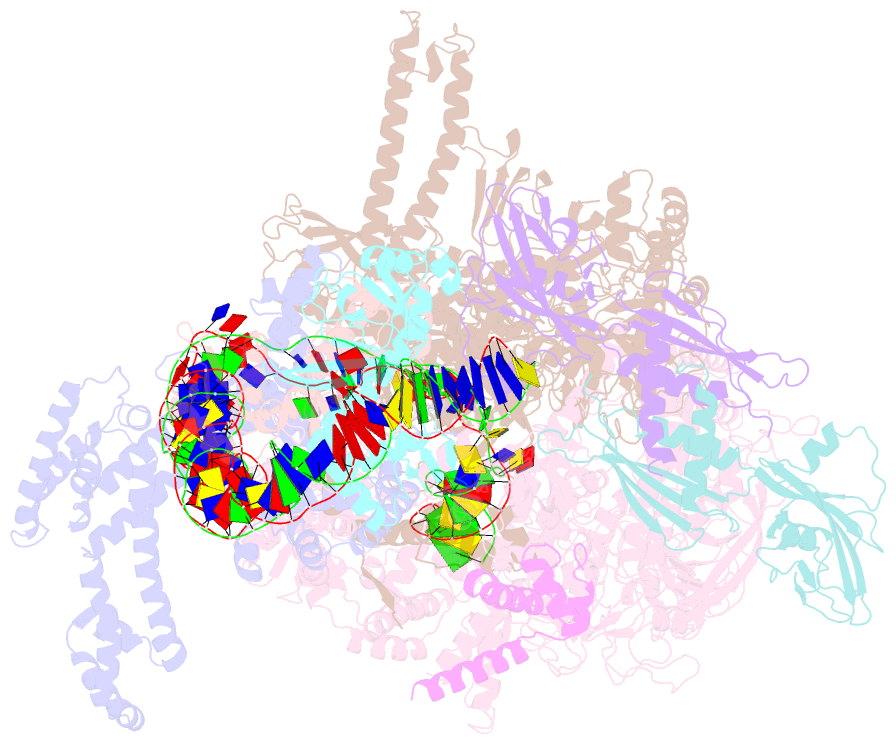
SNAP output for PDB entry 6pb6 [SNAP web server]
Summary information and primary citation
- PDB-id
- 6pb6; SNAP-derived features in text and JSON formats; DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (4.29 Å)
- Summary
- The e. coli class-ii cap-dependent transcription activation complex at the state 2
- Reference
- Shi W, Jiang Y, Deng Y, Dong Z, Liu B (2020): "Visualization of two architectures in class-II CAP-dependent transcription activation." Plos Biol., 18, e3000706. doi: 10.1371/journal.pbio.3000706.
- Abstract