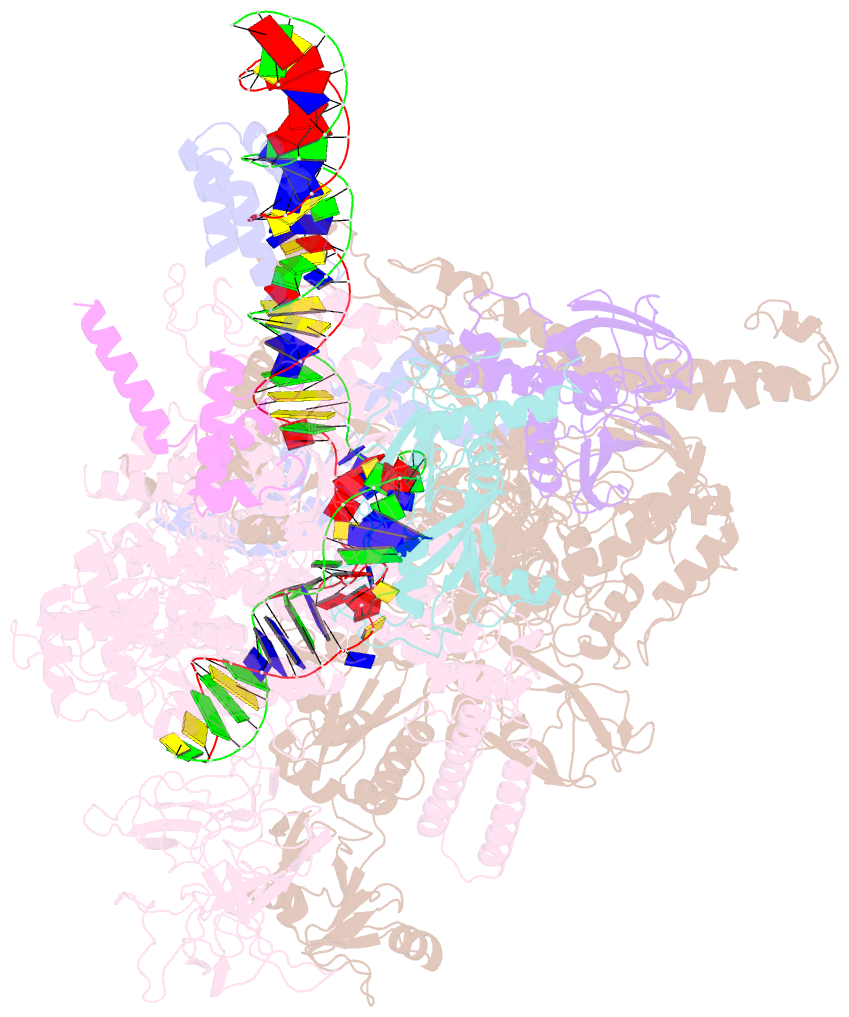
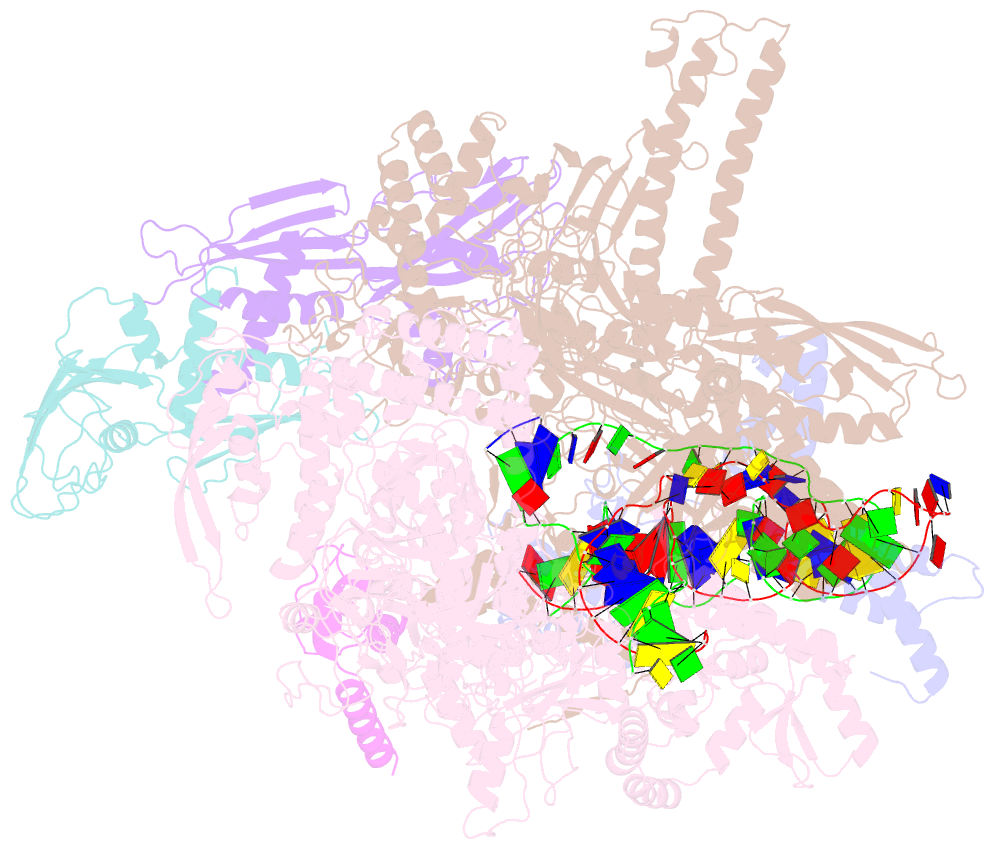
Summary information and primary citation
- PDB-id
- 6pmi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.86 Å)
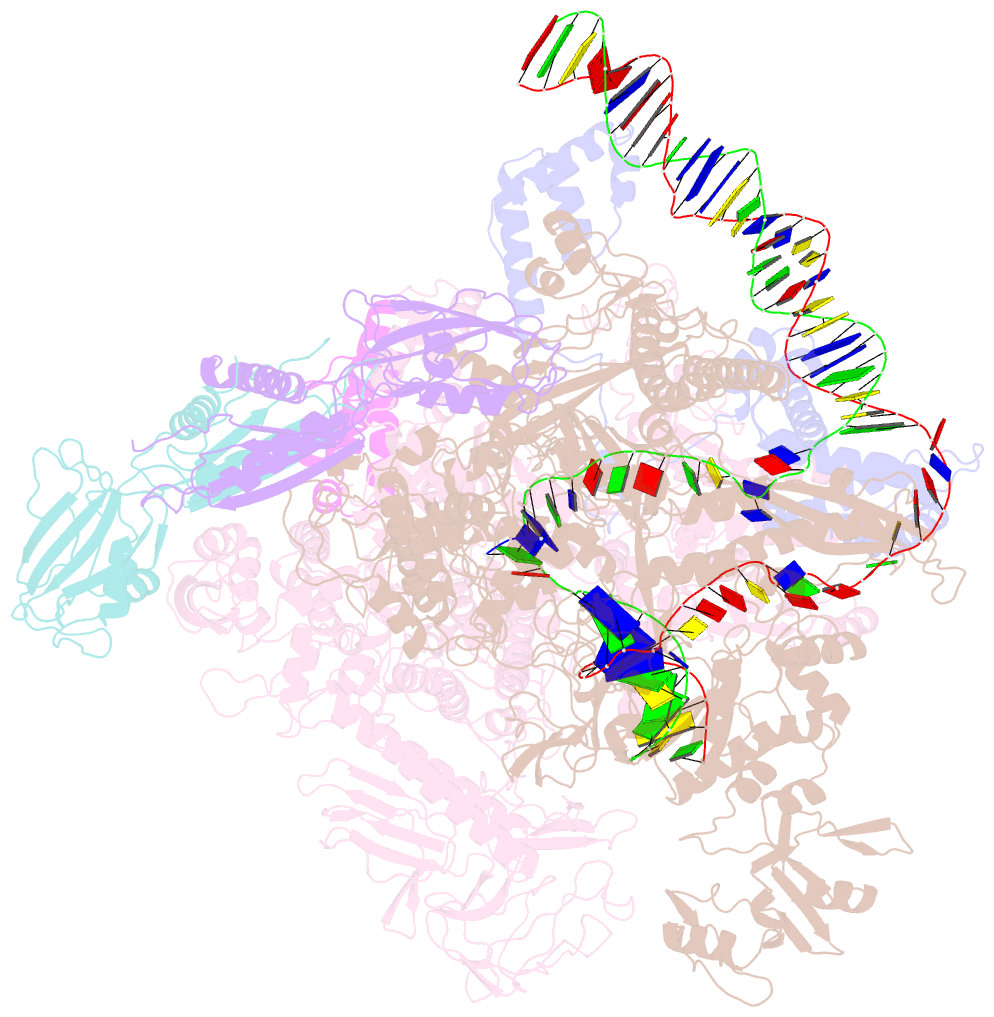
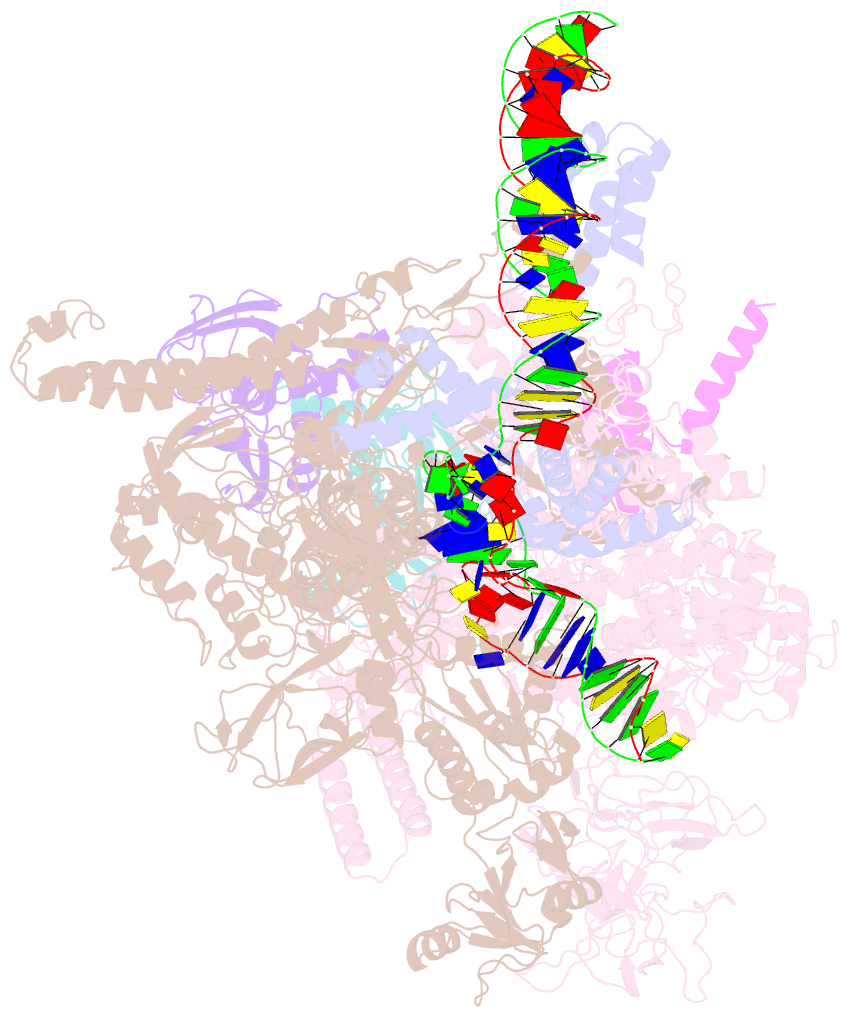
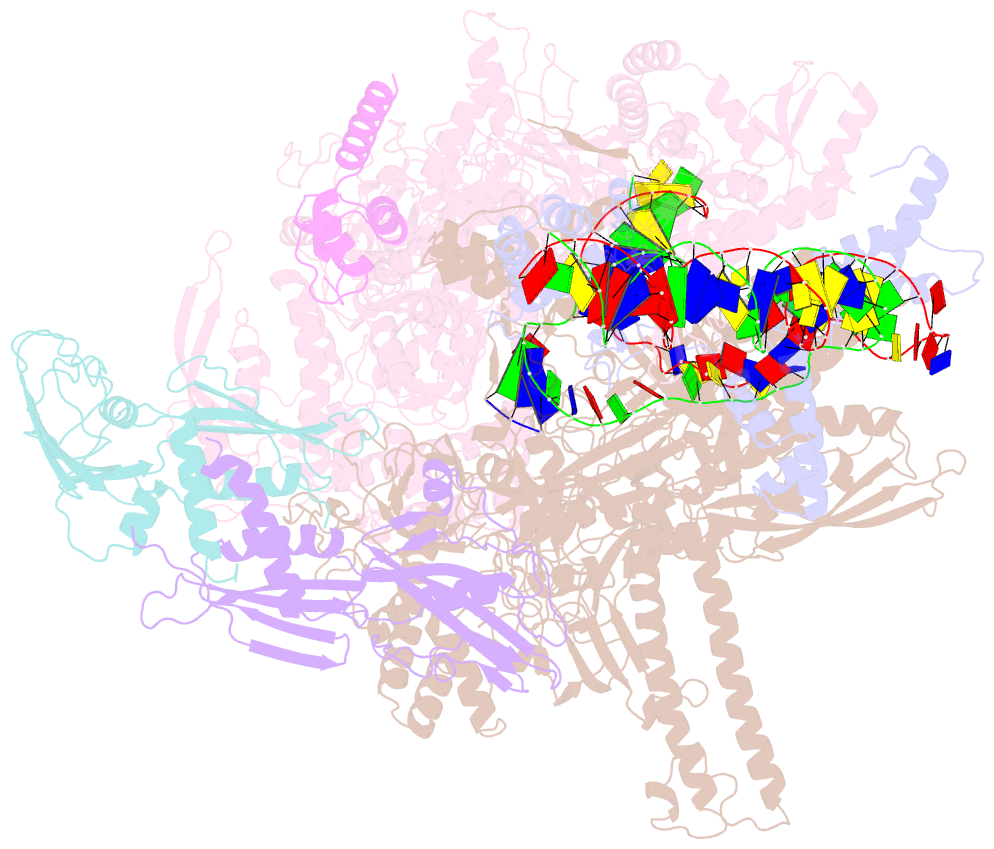
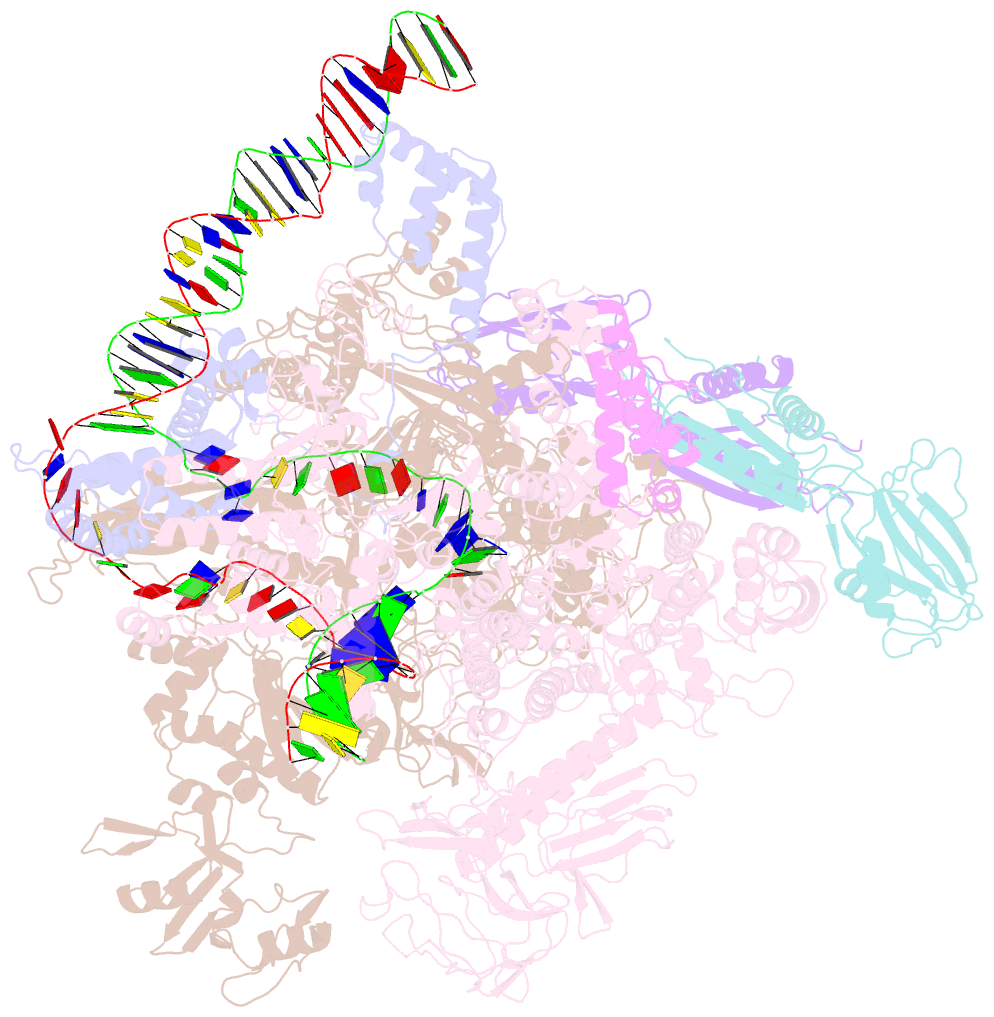
- Summary
- Sigm28-transcription initiation complex with specific promoter at the state 1
- Reference
- Shi W, Zhou W, Zhang B, Huang S, Jiang Y, Schammel A, Hu Y, Liu B (2020): "Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain." Embo J., 39, e104389. doi: 10.15252/embj.2020104389.
- Abstract
- In bacteria, σ28 is the flagella-specific sigma factor that targets RNA polymerase (RNAP) to control the expression of flagella-related genes involving bacterial motility and chemotaxis. However, the structural mechanism of σ28 -dependent promoter recognition remains uncharacterized. Here, we report cryo-EM structures of E. coli σ28 -dependent transcribing complexes on a complete flagella-specific promoter. These structures reveal how σ28 -RNAP recognizes promoter DNA through strong interactions with the -10 element, but weak contacts with the -35 element, to initiate transcription. In addition, we observed a distinct architecture in which the β' zinc-binding domain (ZBD) of RNAP stretches out from its canonical position to interact with the upstream non-template strand. Further in vitro and in vivo assays demonstrate that this interaction has the overall effect of facilitating closed-to-open isomerization of the RNAP-promoter complex by compensating for the weak interaction between σ4 and -35 element. This suggests that ZBD relocation may be a general mechanism employed by σ70 family factors to enhance transcription from promoters with weak σ4/-35 element interactions.