Summary information and primary citation
- PDB-id
- 6ppn; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.91 Å)
- Summary
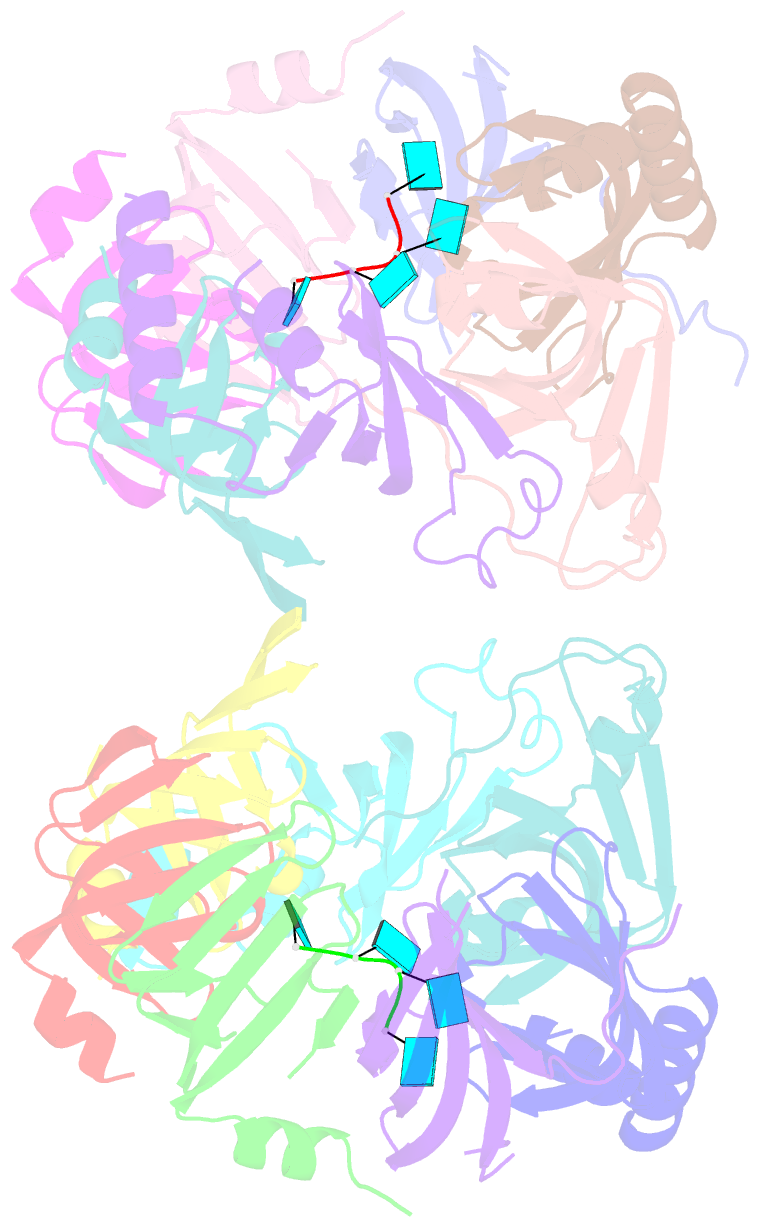
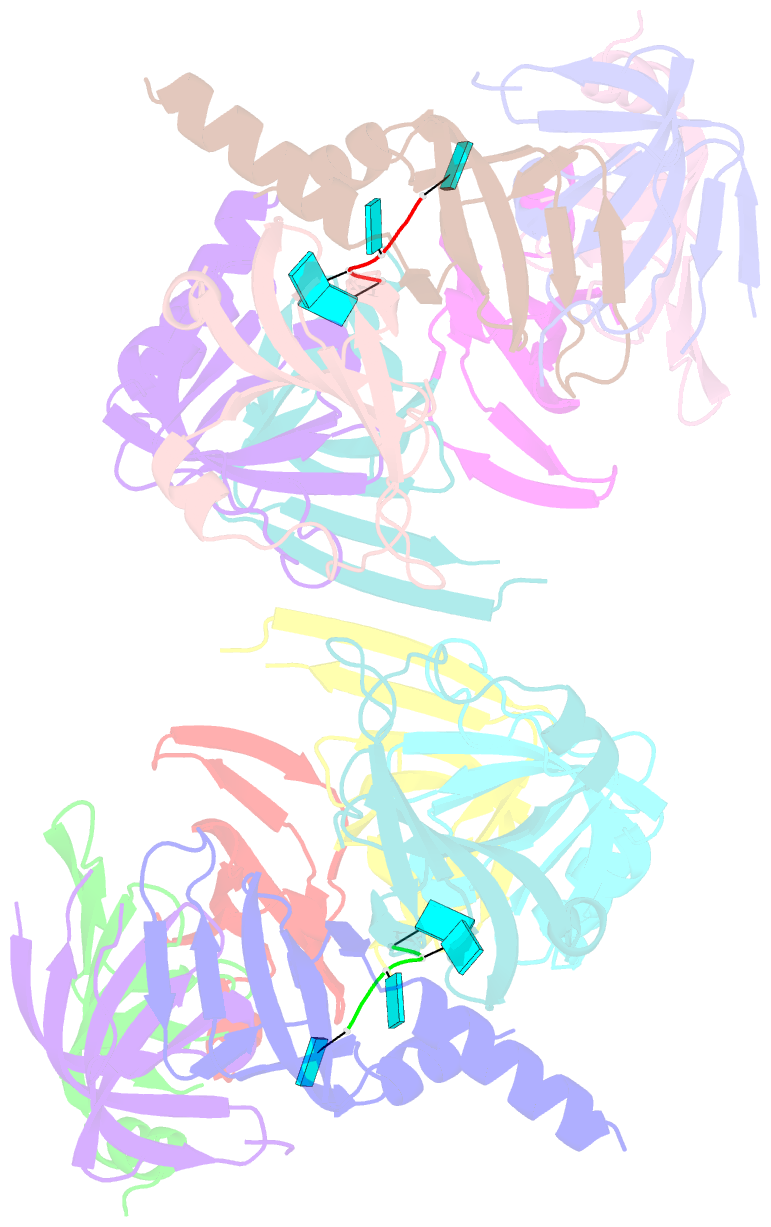
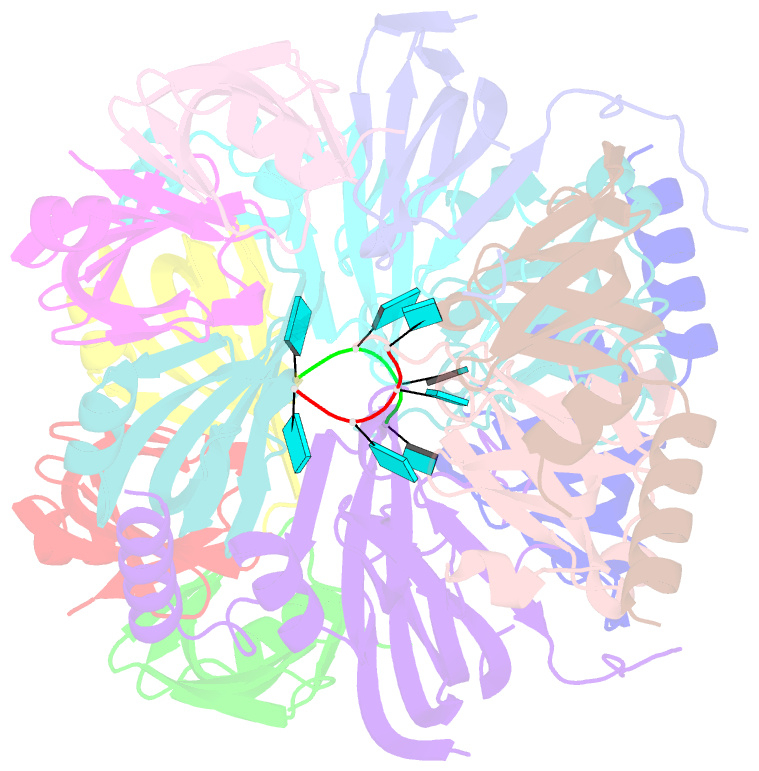
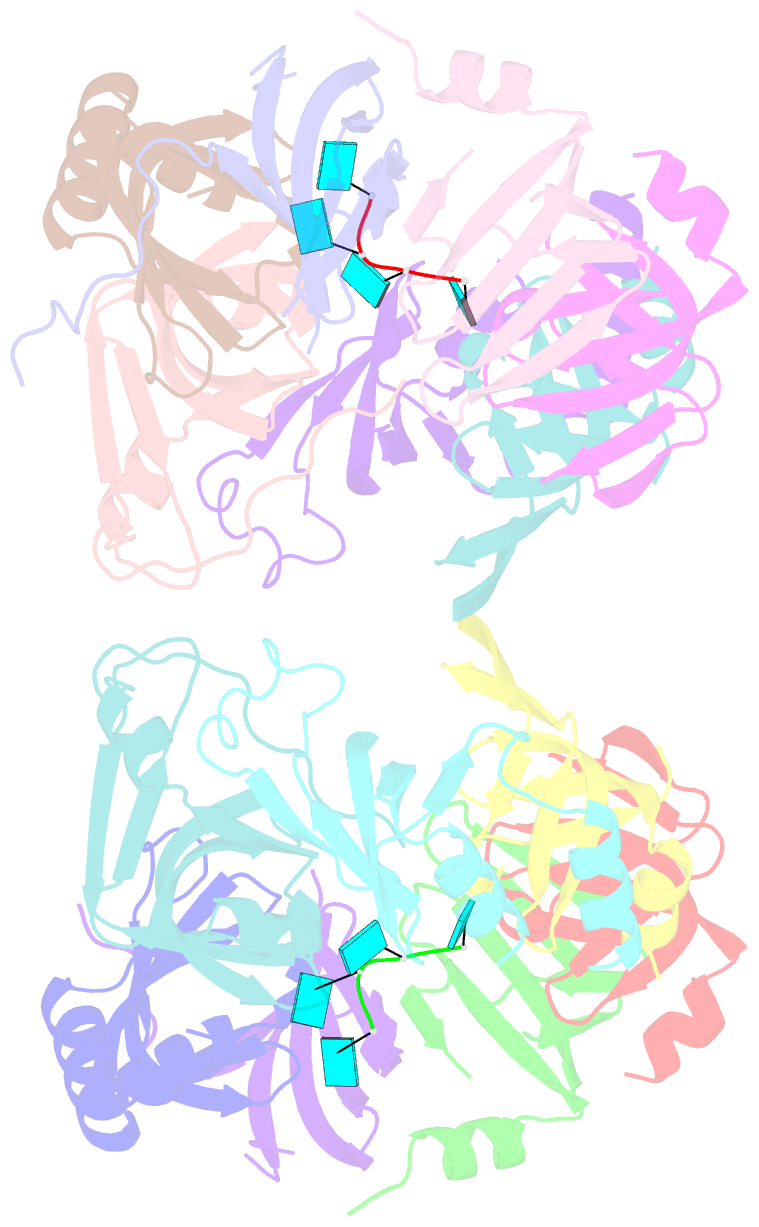
- Structure of s. pombe lsm2-8 with unprocessed u6 snrna
- Reference
- Montemayor EJ, Virta JM, Hayes SM, Nomura Y, Brow DA, Butcher SE (2020): "Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes." Rna, 26, 1400-1413. doi: 10.1261/rna.075879.120.
- Abstract
- Eukaryotes possess eight highly conserved Lsm (like Sm) proteins that assemble into circular, heteroheptameric complexes, bind RNA, and direct a diverse range of biological processes. Among the many essential functions of Lsm proteins, the cytoplasmic Lsm1-7 complex initiates mRNA decay, while the nuclear Lsm2-8 complex acts as a chaperone for U6 spliceosomal RNA. It has been unclear how these complexes perform their distinct functions while differing by only one out of seven subunits. Here, we elucidate the molecular basis for Lsm-RNA recognition and present four high-resolution structures of Lsm complexes bound to RNAs. The structures of Lsm2-8 bound to RNA identify the unique 2',3' cyclic phosphate end of U6 as a prime determinant of specificity. In contrast, the Lsm1-7 complex strongly discriminates against cyclic phosphates and tightly binds to oligouridylate tracts with terminal purines. Lsm5 uniquely recognizes purine bases, explaining its divergent sequence relative to other Lsm subunits. Lsm1-7 loads onto RNA from the 3' end and removal of the Lsm1 carboxy-terminal region allows Lsm1-7 to scan along RNA, suggesting a gated mechanism for accessing internal binding sites. These data reveal the molecular basis for RNA binding by Lsm proteins, a fundamental step in the formation of molecular assemblies that are central to eukaryotic mRNA metabolism.