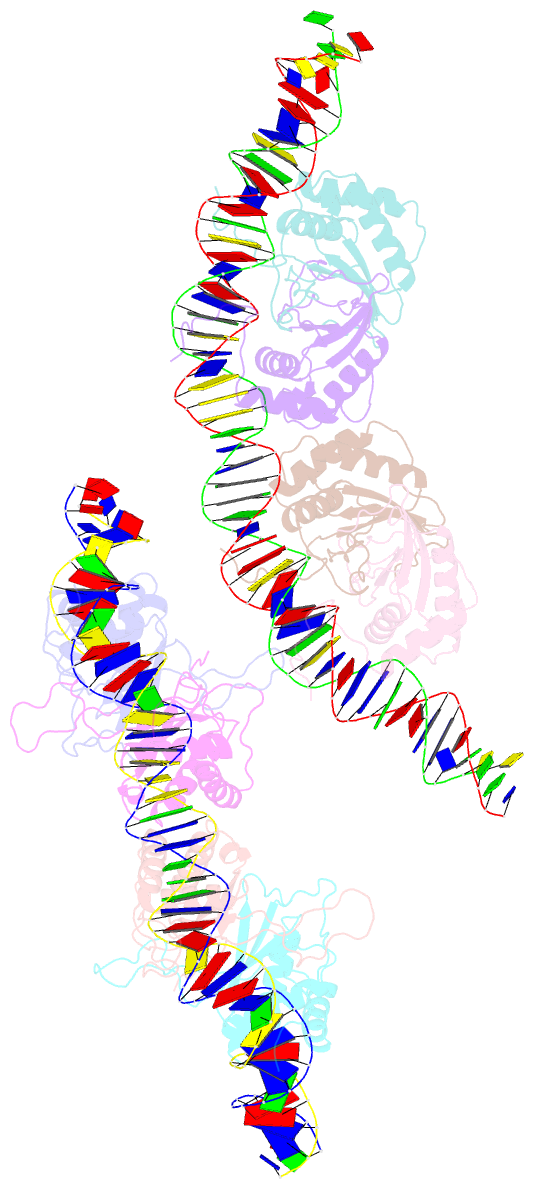
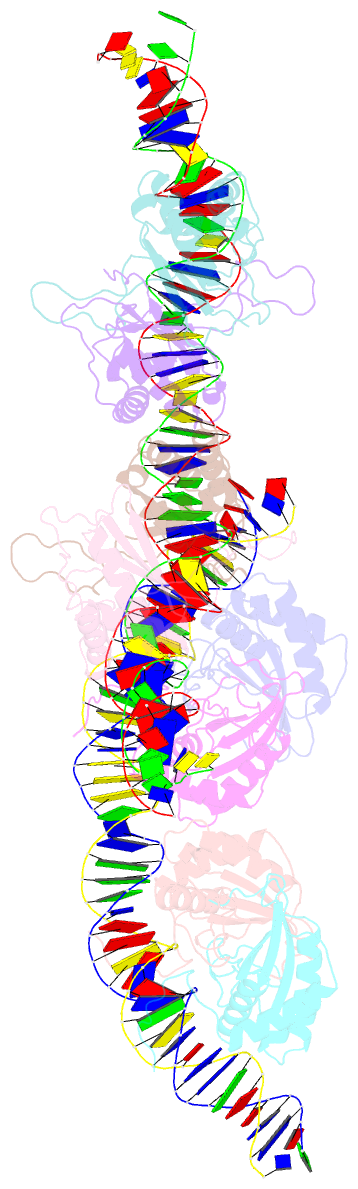
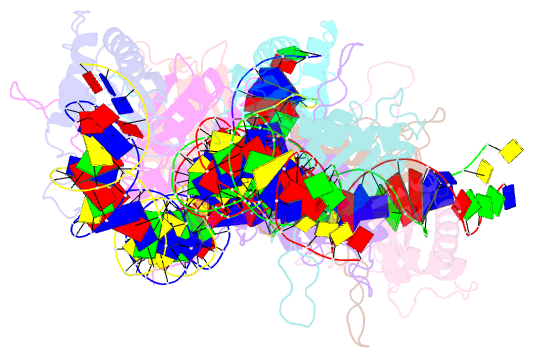
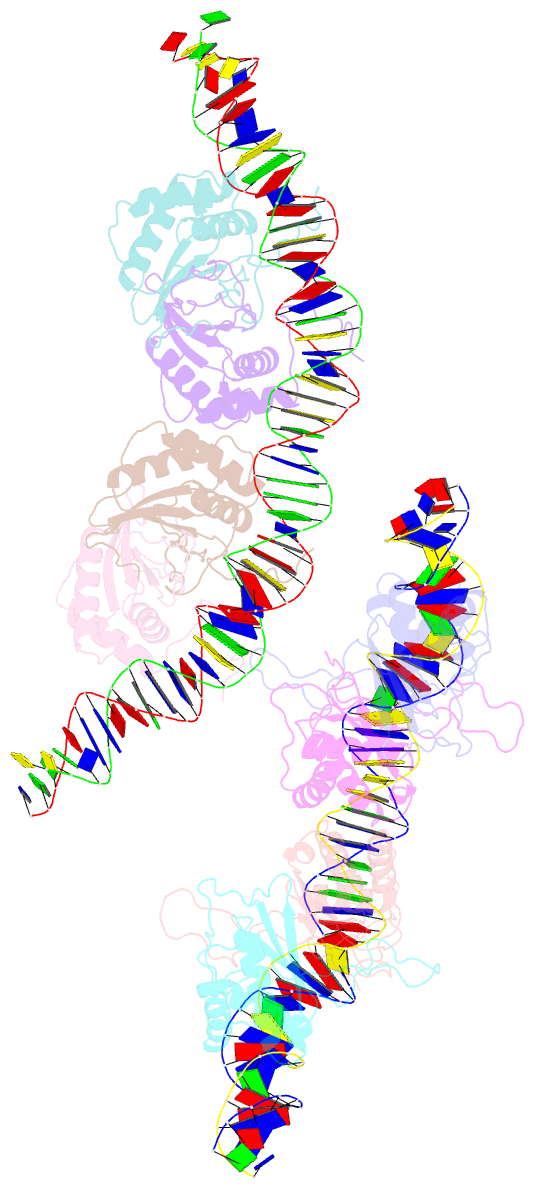
Summary information and primary citation
- PDB-id
- 6pw2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- X-ray (3.01 Å)
- Summary
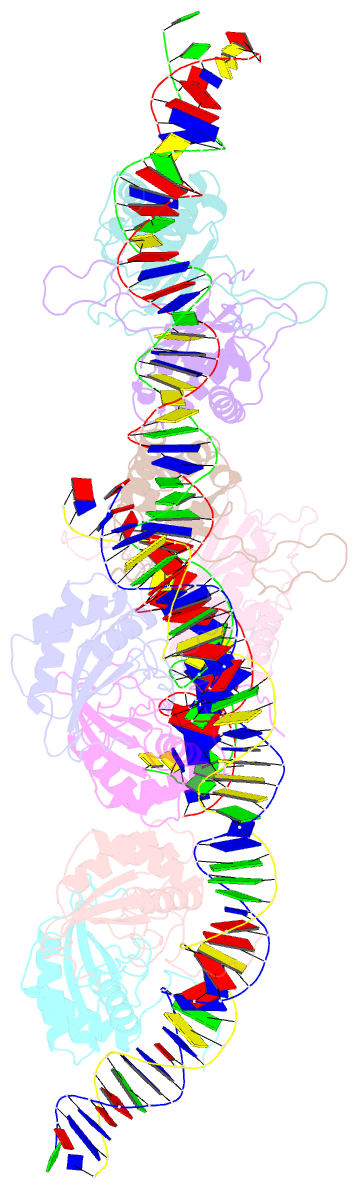
- Structural basis for cooperative binding of ebna1 to the epstein-barr virus dyad symmetry minimal origin of replication
- Reference
- Malecka KA, Dheekollu J, Deakyne JS, Wiedmer A, Ramirez UD, Lieberman PM, Messick TE (2019): "Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication." J.Virol., 93. doi: 10.1128/JVI.00487-19.
- Abstract
- Epstein-Barr virus is associated with several human malignancies, including nasopharyngeal carcinoma, gastric cancer, and lymphoma. Latently infected cells carry a circularized EBV episome where the origin of replication (oriP) is comprised of two elements: the family of repeats (FR) and dyad symmetry (DS). The viral protein Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) binds to FR and DS to promote EBV episome maintenance and DNA replication during latent infection in proliferating cells. EBNA1 binding to the DS constitutes a minimal origin of DNA replication. Here we report the crystal structure of two EBNA1 DNA-binding domain dimers bound to a DS half-site. This structure shows that the DNA is smoothly bent, allowing for stabilizing interactions between the dimers. The dimer-dimer interface requires an intricate hydrogen bonding network involving residues R491 and D581. When this interface is disrupted, we note loss of stable dimer-dimer complex formation on the DNA, compromised oriP-containing plasmid replication in cells, and impaired recruitment of the MCM3 complex to the oriP Surface conservation analysis reveals that these residues are part of a larger conserved surface that may be critical for recruitment of replication machinery to the oriP Our results reveal a new region of EBNA1 critical for its activity and one that may be exploited by targeted small molecules to treat EBV-associated disease.