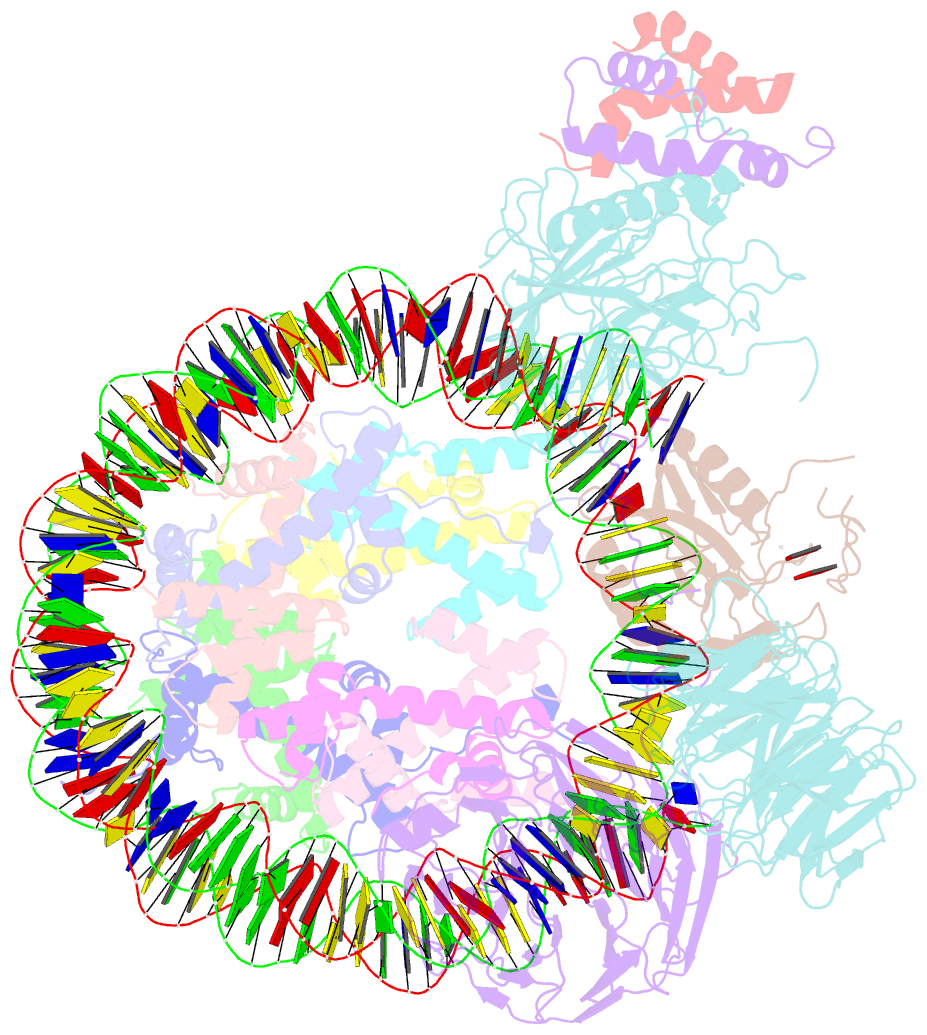
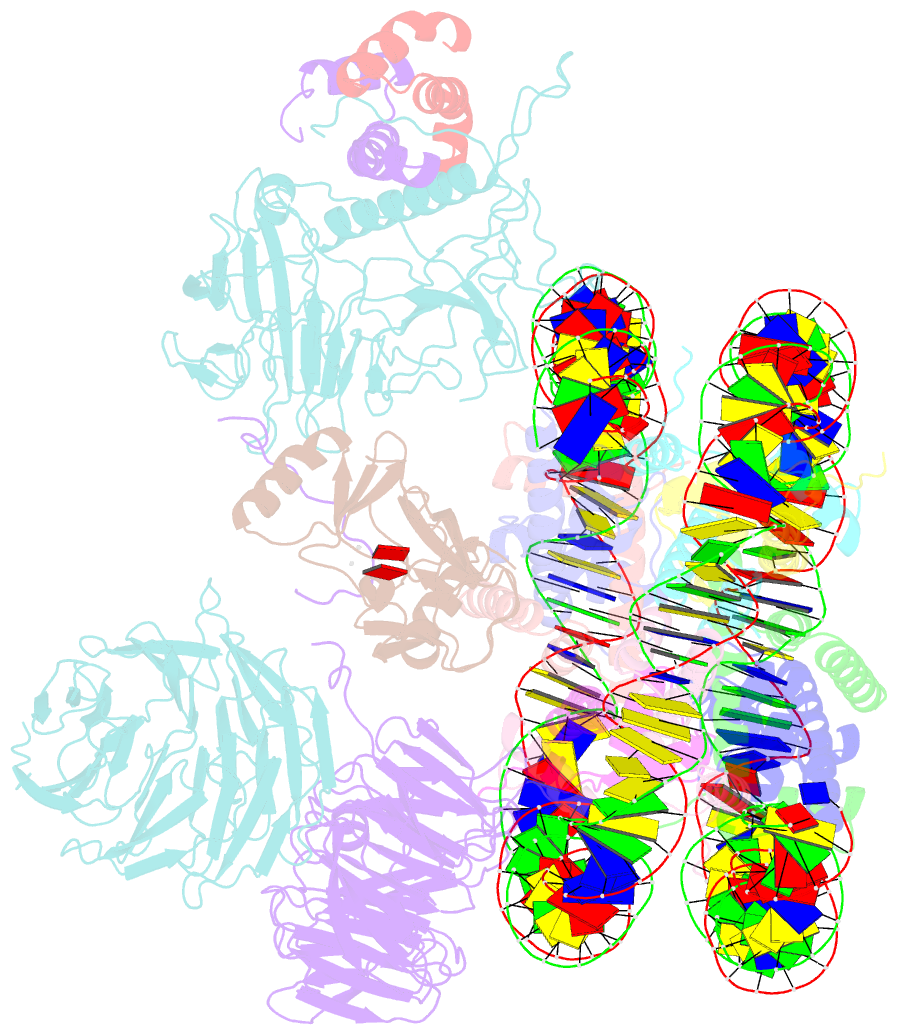
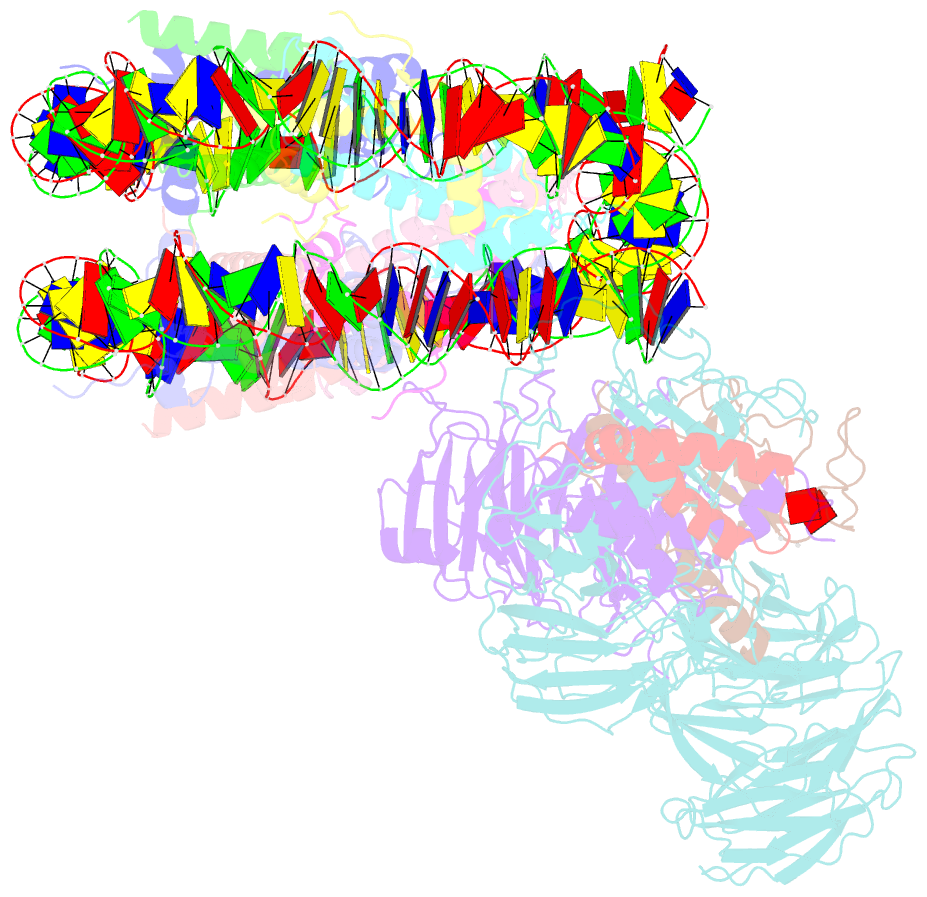
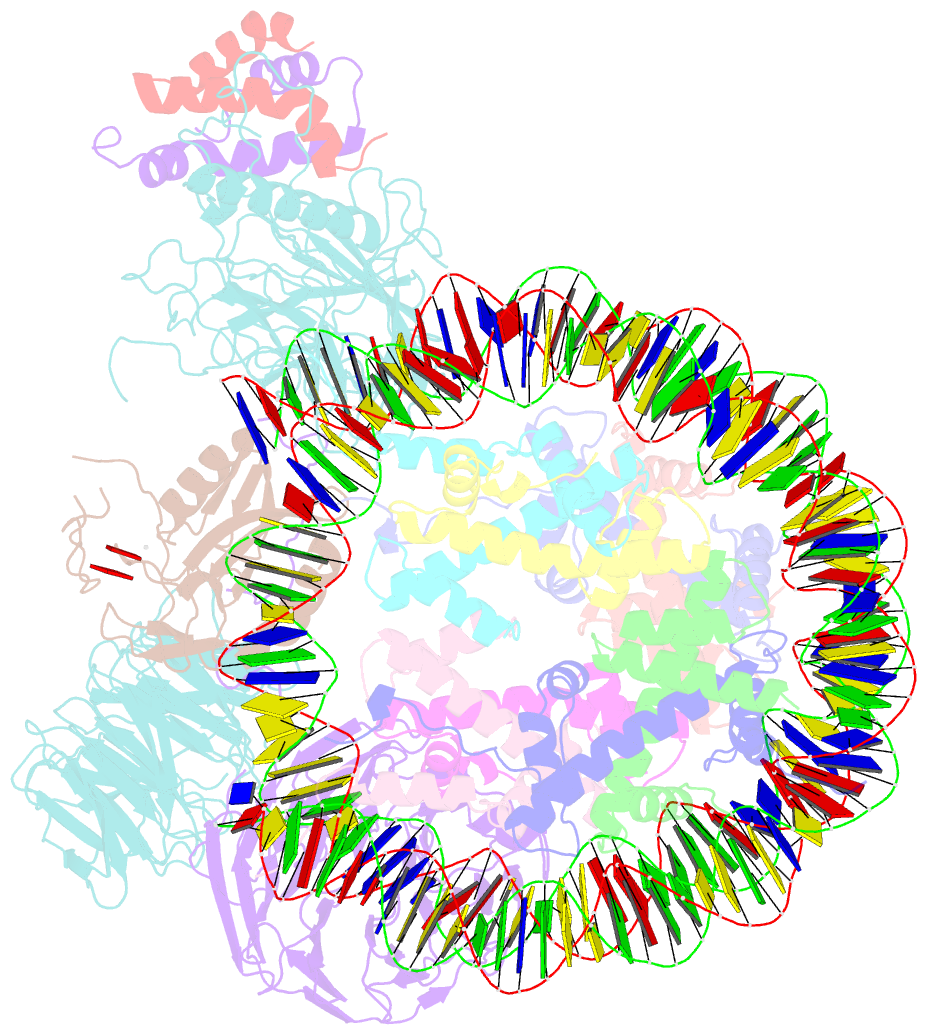
Summary information and primary citation
- PDB-id
- 6pwv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- histone binding-DNA binding-DNA
- Method
- cryo-EM (6.2 Å)
- Summary
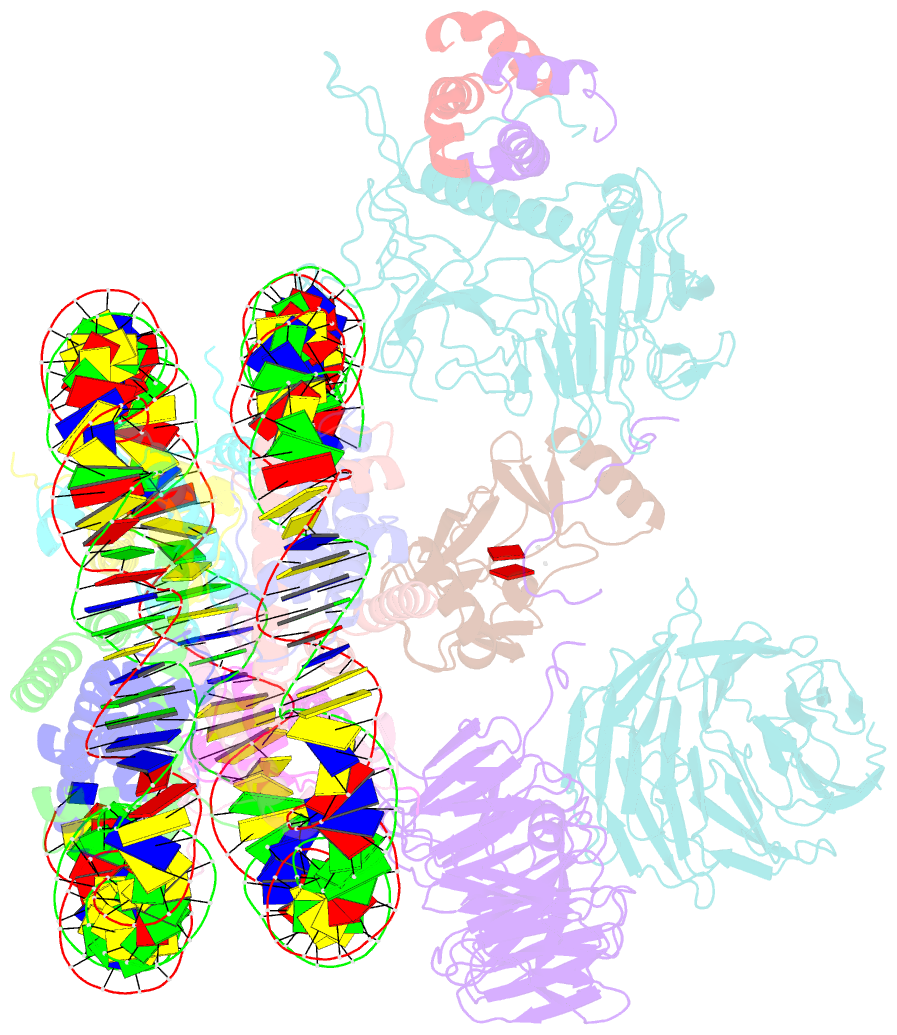
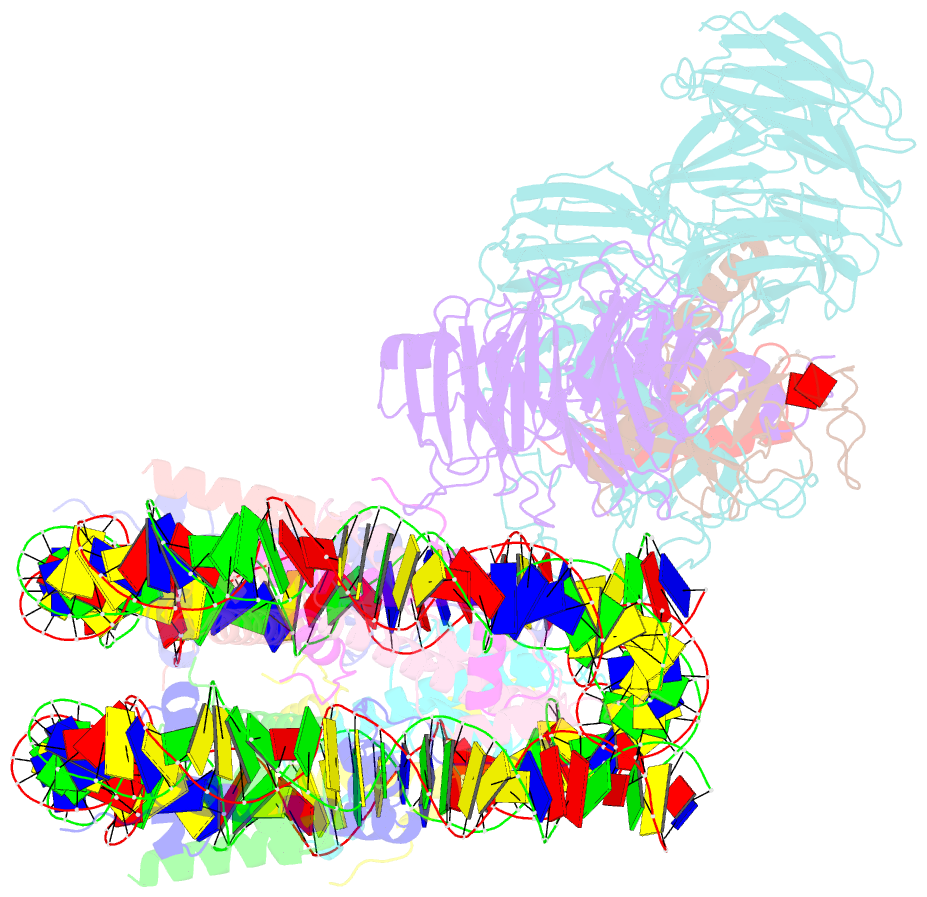
- cryo-EM structure of mll1 core complex bound to the nucleosome
- Reference
- Park SH, Ayoub A, Lee YT, Xu J, Kim H, Zheng W, Zhang B, Sha L, An S, Zhang Y, Cianfrocco MA, Su M, Dou Y, Cho US (2019): "Cryo-EM structure of the human MLL1 core complex bound to the nucleosome." Nat Commun, 10, 5540. doi: 10.1038/s41467-019-13550-2.
- Abstract
- Mixed lineage leukemia (MLL) family histone methyltransferases are enzymes that deposit histone H3 Lys4 (K4) mono-/di-/tri-methylation and regulate gene expression in mammals. Despite extensive structural and biochemical studies, the molecular mechanisms whereby the MLL complexes recognize histone H3K4 within nucleosome core particles (NCPs) remain unclear. Here we report the single-particle cryo-electron microscopy (cryo-EM) structure of the NCP-bound human MLL1 core complex. We show that the MLL1 core complex anchors to the NCP via the conserved RbBP5 and ASH2L, which interact extensively with nucleosomal DNA and the surface close to the N-terminal tail of histone H4. Concurrent interactions of RbBP5 and ASH2L with the NCP uniquely align the catalytic MLL1SET domain at the nucleosome dyad, thereby facilitating symmetrical access to both H3K4 substrates within the NCP. Our study sheds light on how the MLL1 complex engages chromatin and how chromatin binding promotes MLL1 tri-methylation activity.