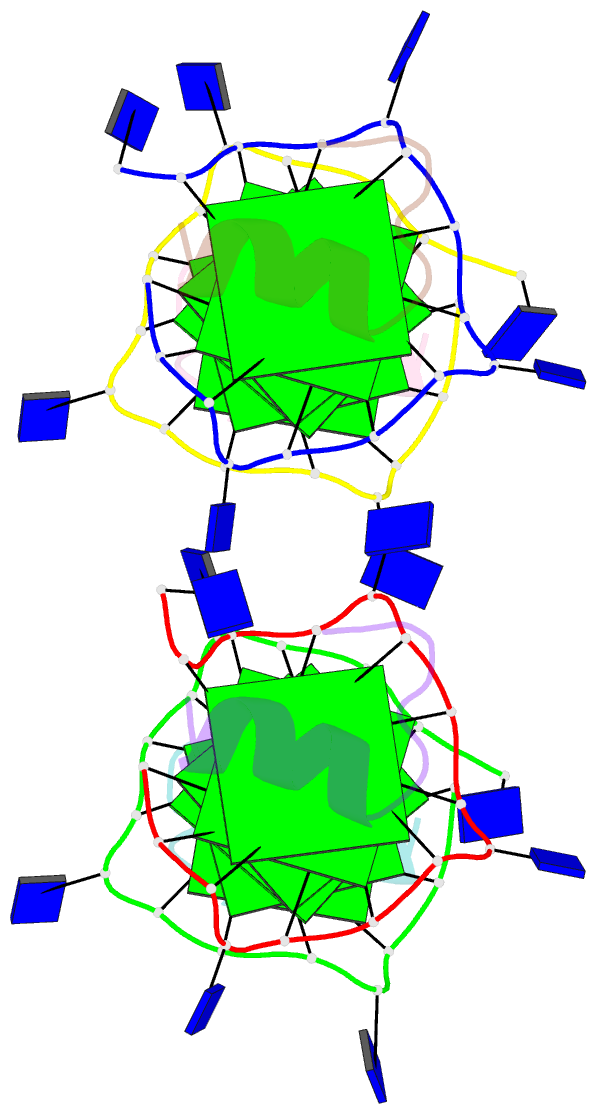
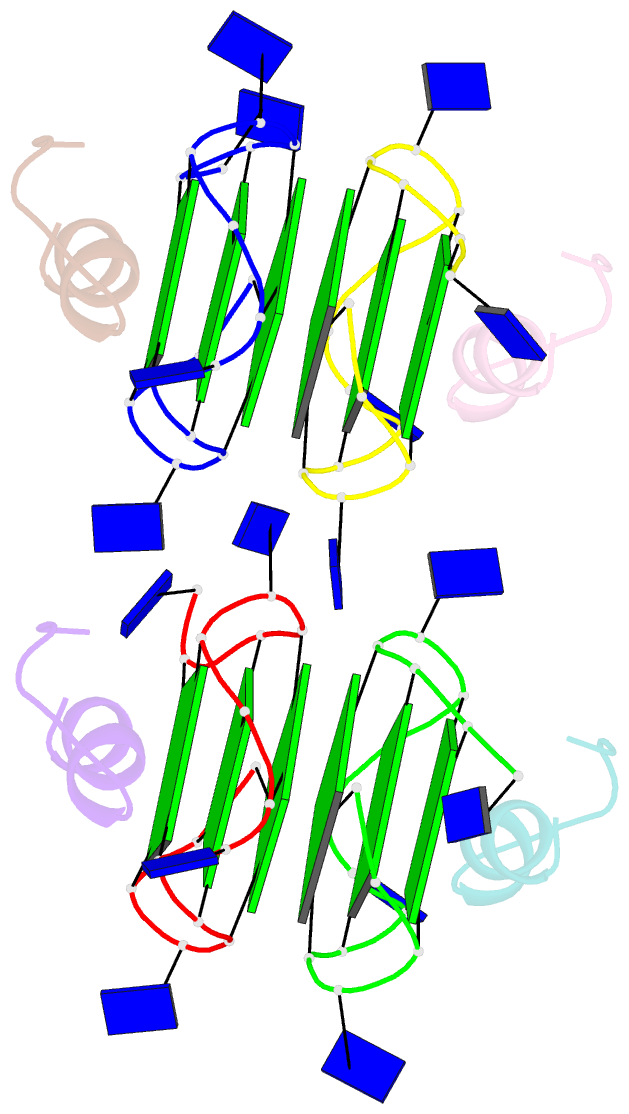
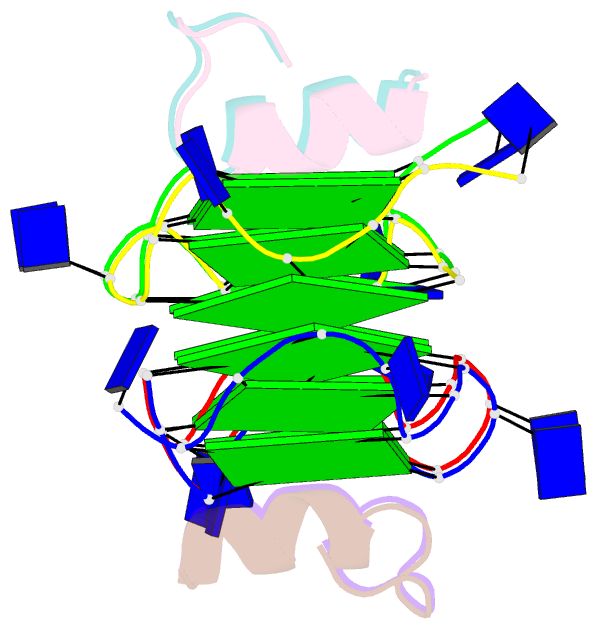
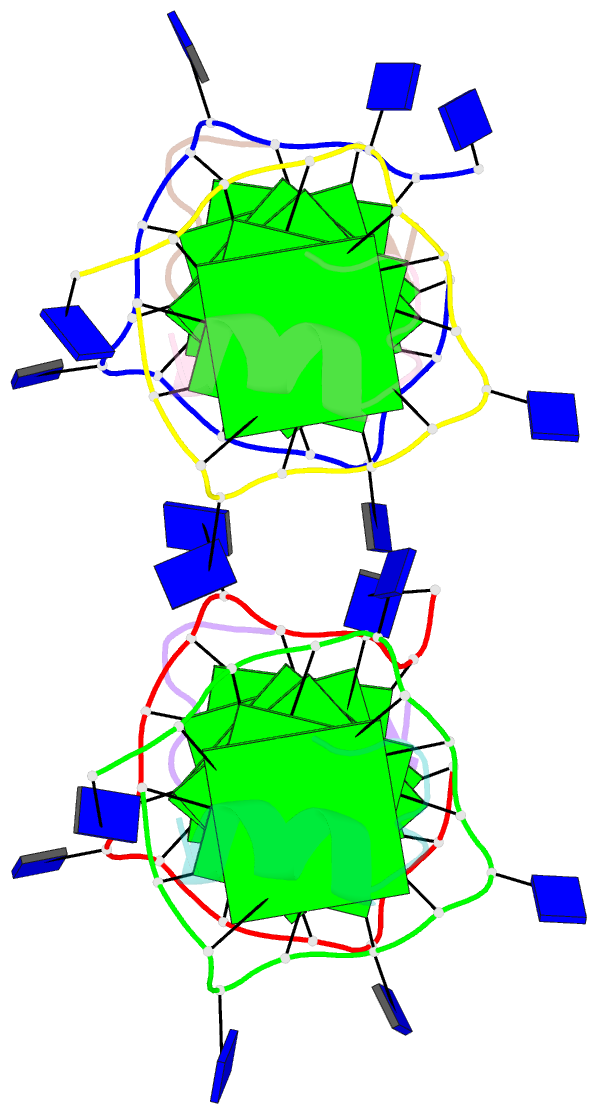
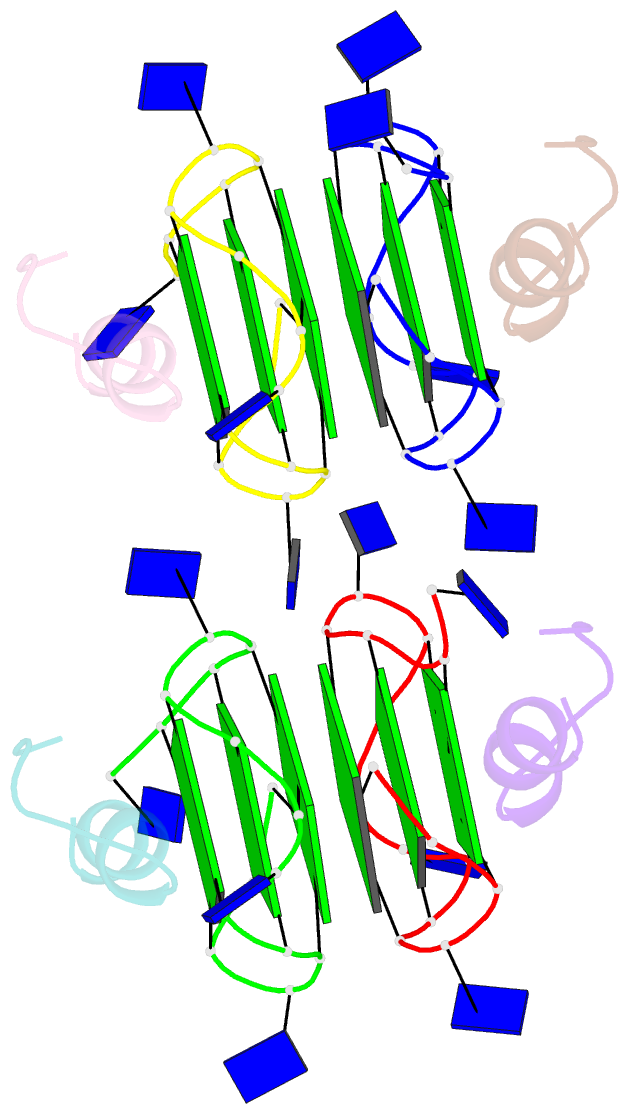
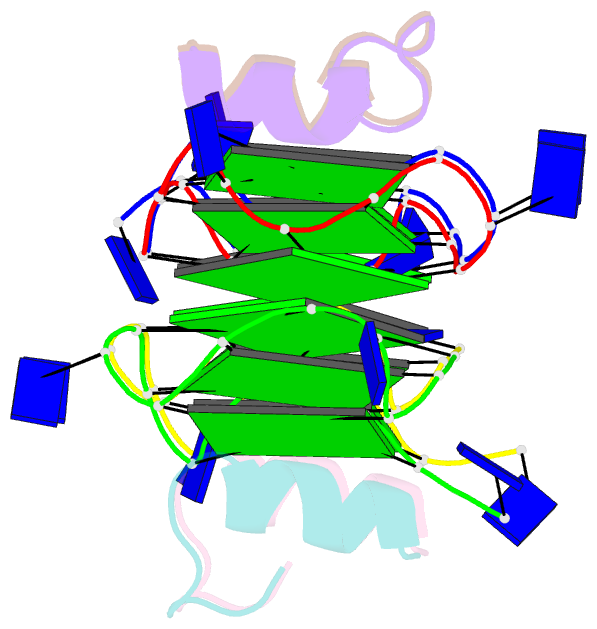
Summary information and primary citation
- PDB-id
- 6q6r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (1.5 Å)
- Summary
- Recognition of different base tetrads by rhau: x-ray crystal structure of g4 recognition motif bound to the 3-end tetrad of a DNA g-quadruplex
- Reference
- Heddi B, Cheong VV, Schmitt E, Mechulam Y, Phan AT (2020): "Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex." J.Struct.Biol., 209, 107399. doi: 10.1016/j.jsb.2019.10.001.
- Abstract
- G-quadruplexes (G4) are secondary structures of nucleic acids that can form in cells and have diverse biological functions. Several biologically important proteins interact with G-quadruplexes, of which RHAU (or DHX36) - a helicase from the DEAH-box superfamily, was shown to bind and unwind G-quadruplexes efficiently. We report a X-ray co-crystal structure at 1.5 Å resolution of an N-terminal fragment of RHAU bound to an exposed tetrad of a parallel-stranded G-quadruplex. The RHAU peptide folds into an L-shaped α-helix, and binds to a G-quadruplex through π-stacking and electrostatic interactions. X-ray crystal structure of our complex identified key amino acid residues important for G-quadruplex-peptide binding interaction at the 3'-end G•G•G•G tetrad. Together with previous solution and crystal structures of RHAU bound to the 5'-end G•G•G•G and G•G•A•T tetrads, our crystal structure highlights the occurrence of a robust G-quadruplex recognition motif within RHAU that can adapt to different accessible tetrads.