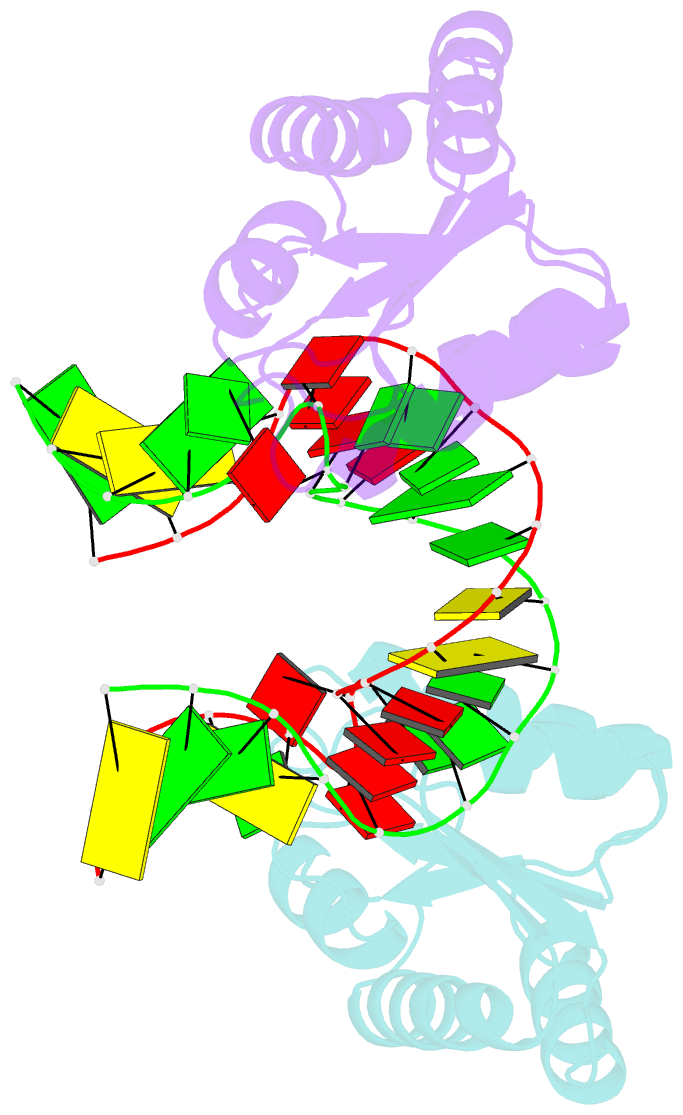
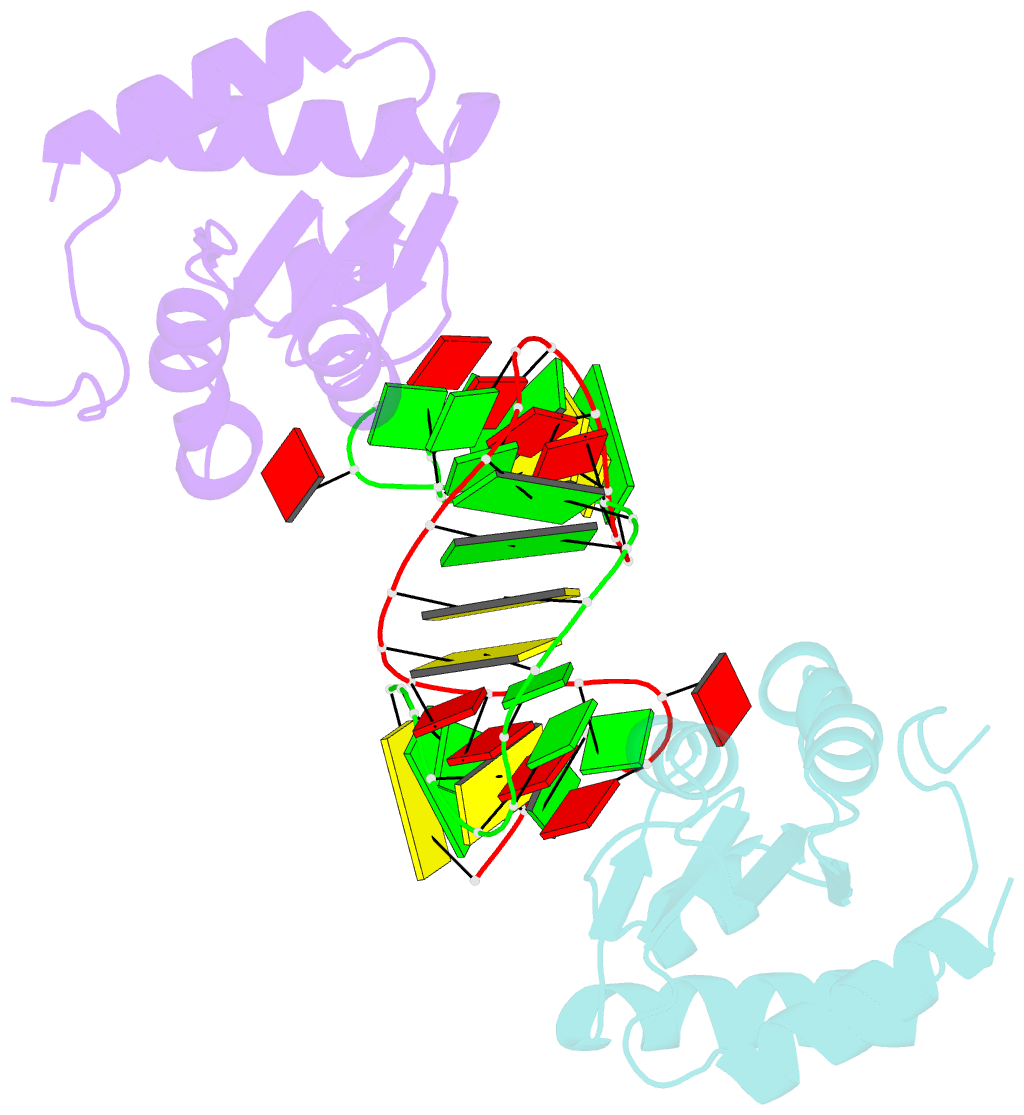
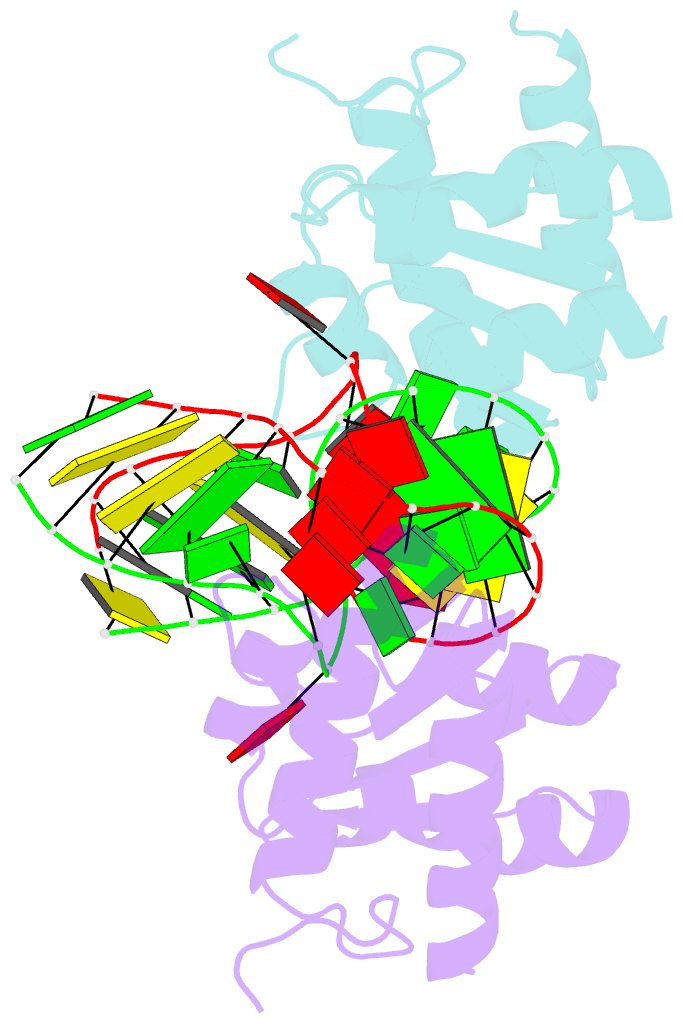
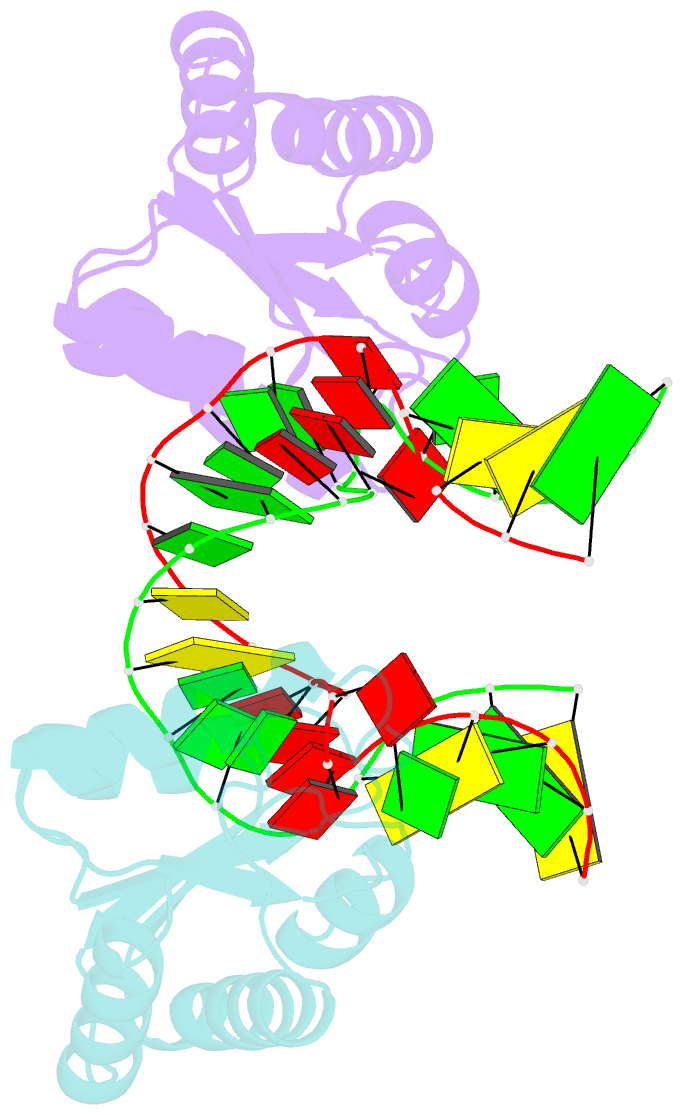
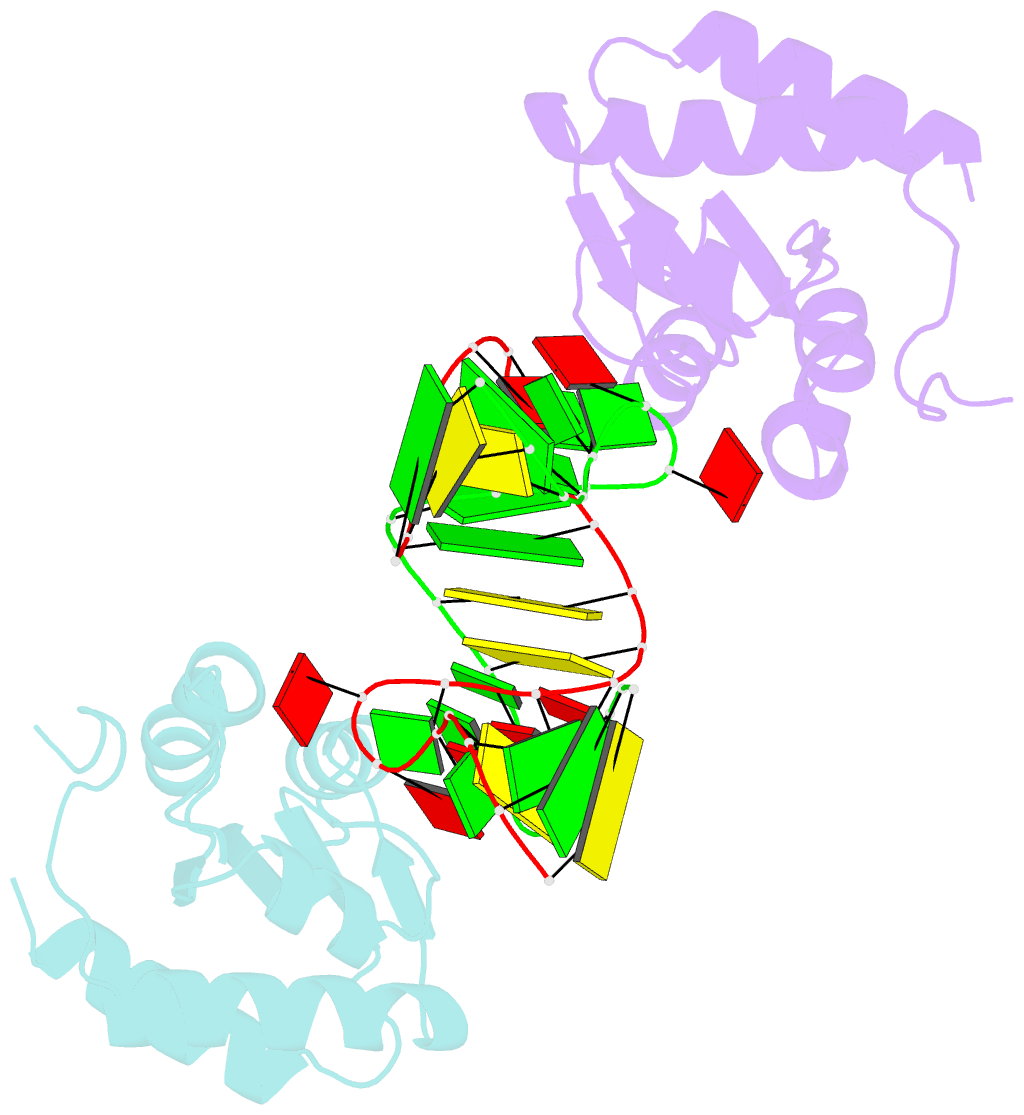
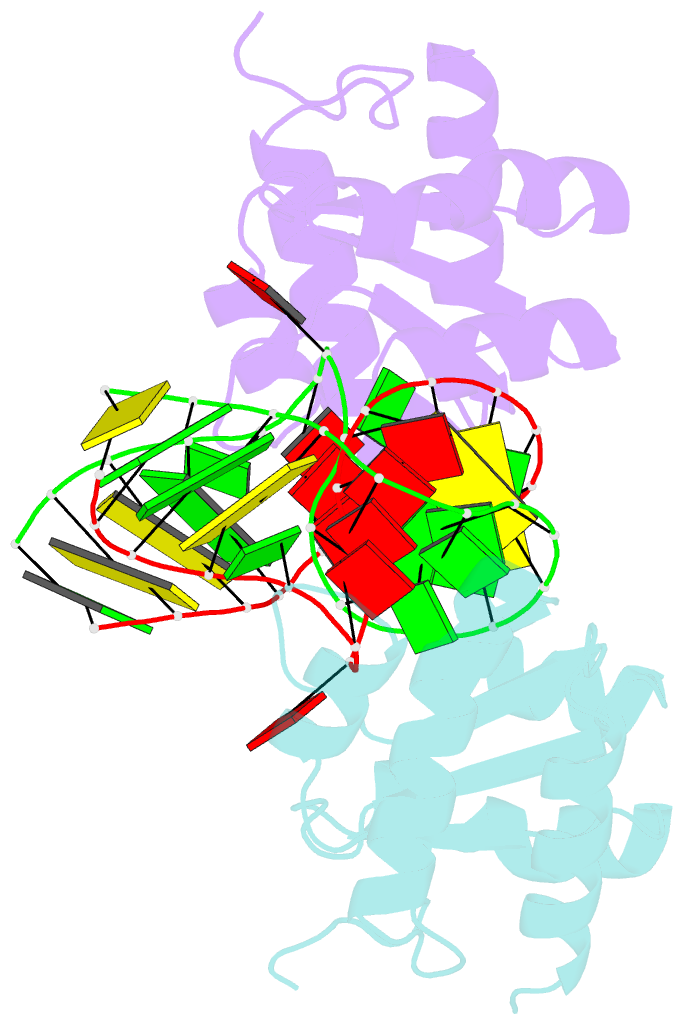
Summary information and primary citation
- PDB-id
- 6q8u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA
- Method
- X-ray (1.99 Å)
- Summary
- Structure of the standard kink turn hmkt-7 variant a2bm6a bound with afl7ae protein
- Reference
- Ashraf S, Huang L, Lilley DMJ (2019): "Effect of methylation of adenine N6on kink turn structure depends on location." Rna Biol., 16, 1377-1385. doi: 10.1080/15476286.2019.1630797.
- Abstract
- N6-methyladenine is the most common covalent modification in cellular RNA species, with demonstrated functional consequences. At the molecular level this methylation could alter local RNA structure, and/or modulate the binding of specific proteins. We have previously shown that trans-Hoogsteen-sugar (sheared) A:G base pairs can be completely disrupted by methylation, and that this occurs in a sub-set ofD/D k-turn structures. In this work we have investigated to what extent sequence context affects the severity with which inclusion of N6-methyladenine into different A:G base pairs of a standard k-turn affects RNA folding and L7Ae protein binding. We find that local sequence has a major influence, ranging from complete absence of folding and protein binding to a relatively mild effect. We have determined the crystal structure of one of these species both free and protein-bound, showing the environment of the methyl group and the way the modification is accommodated into the k-turn structure.