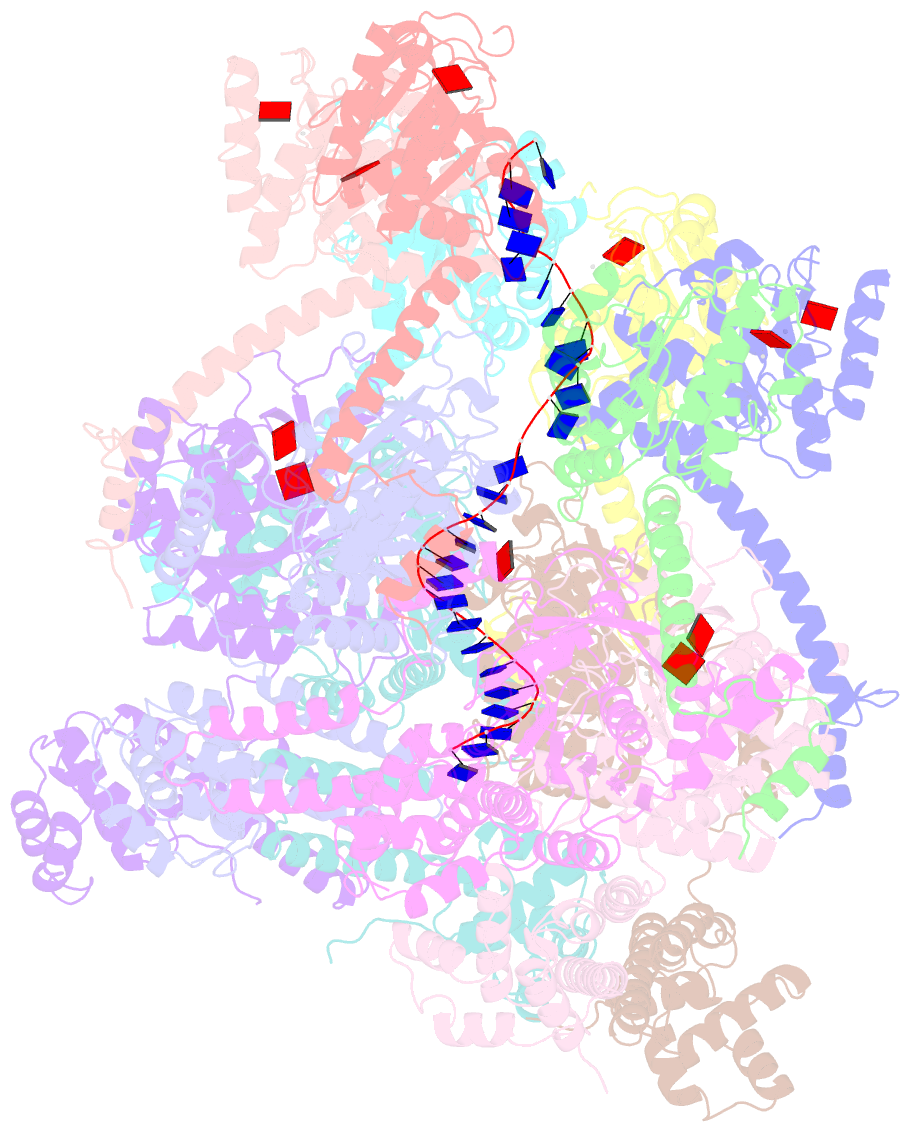
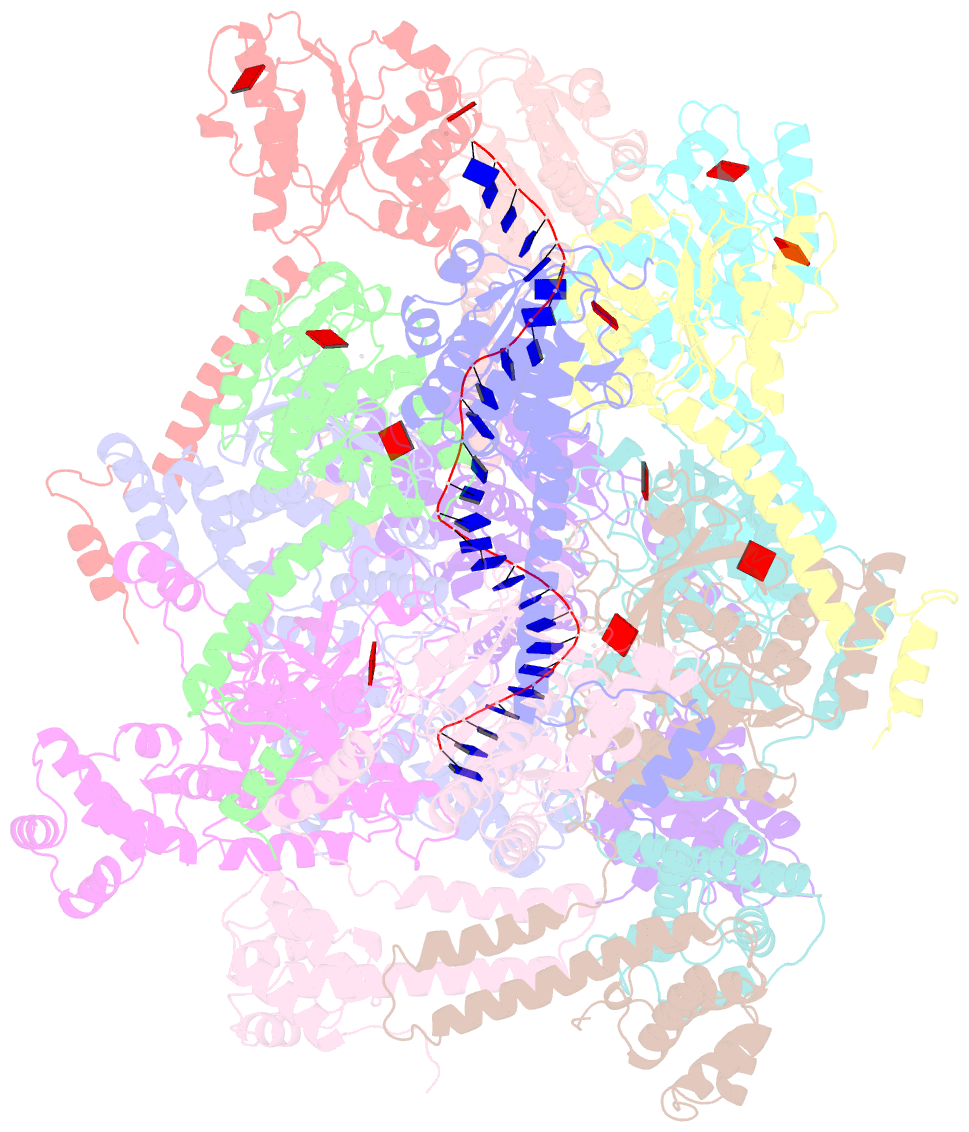
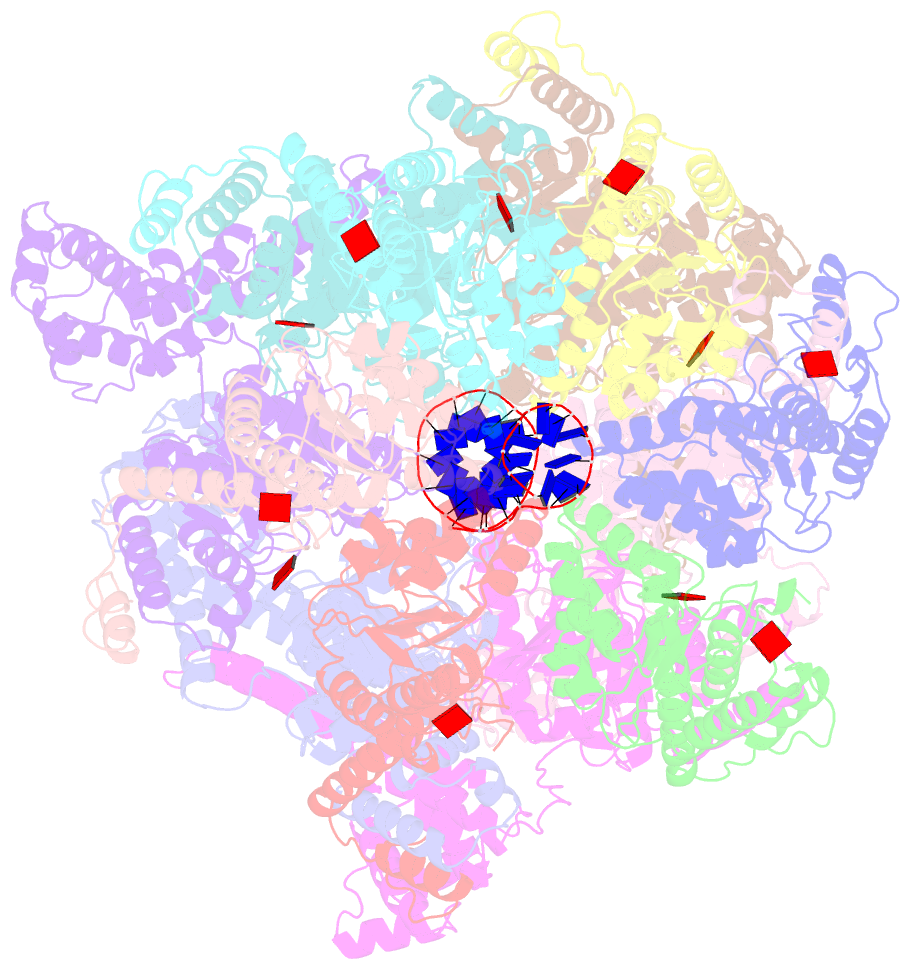
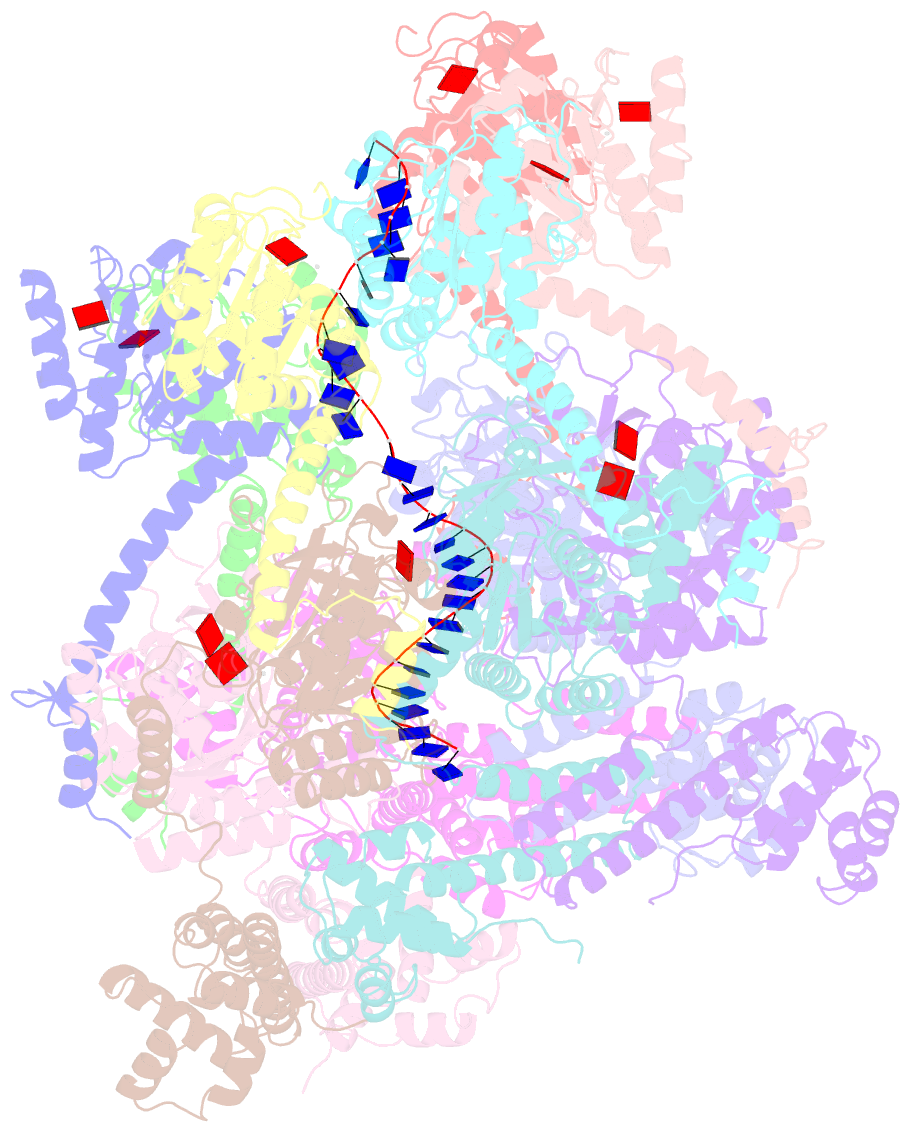
Summary information and primary citation
- PDB-id
- 6qem; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- cryo-EM (3.4 Å)
- Summary
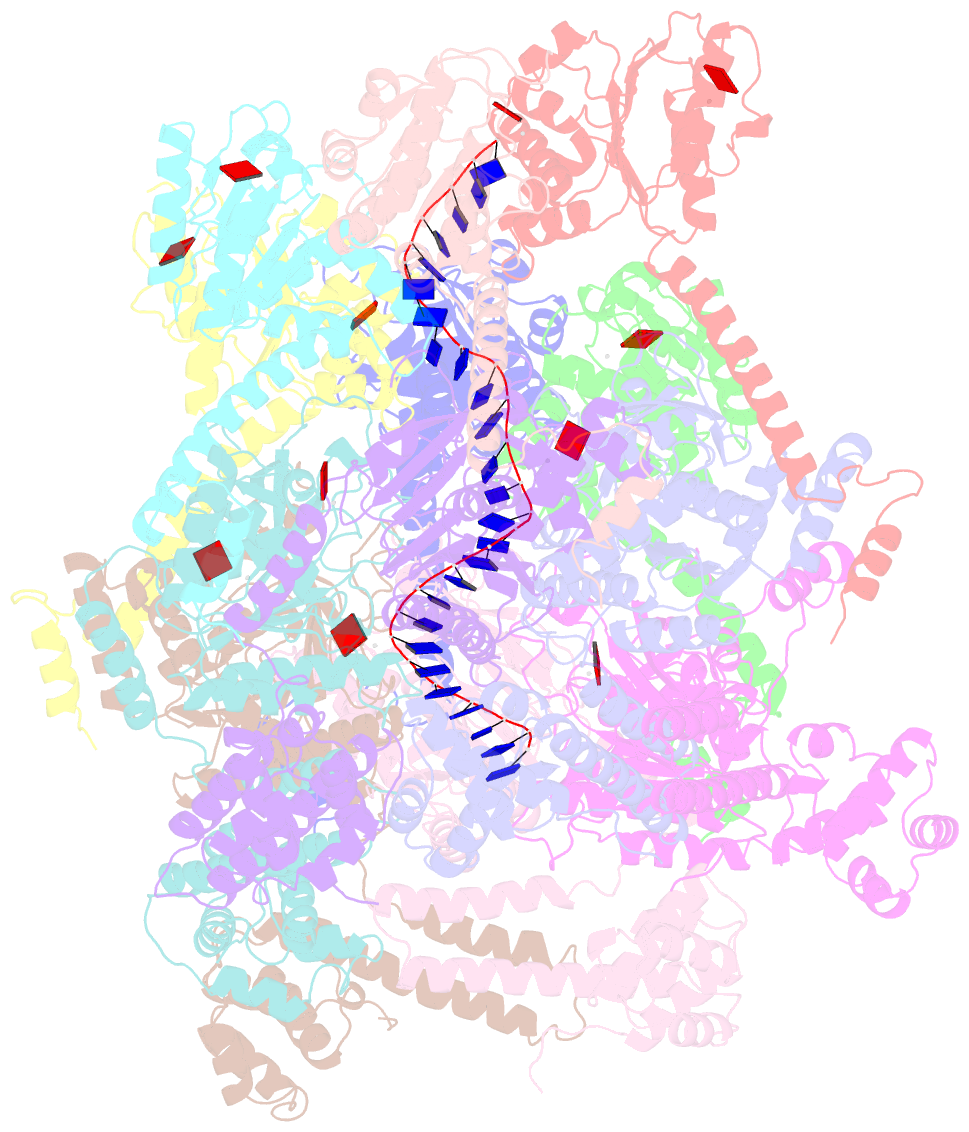
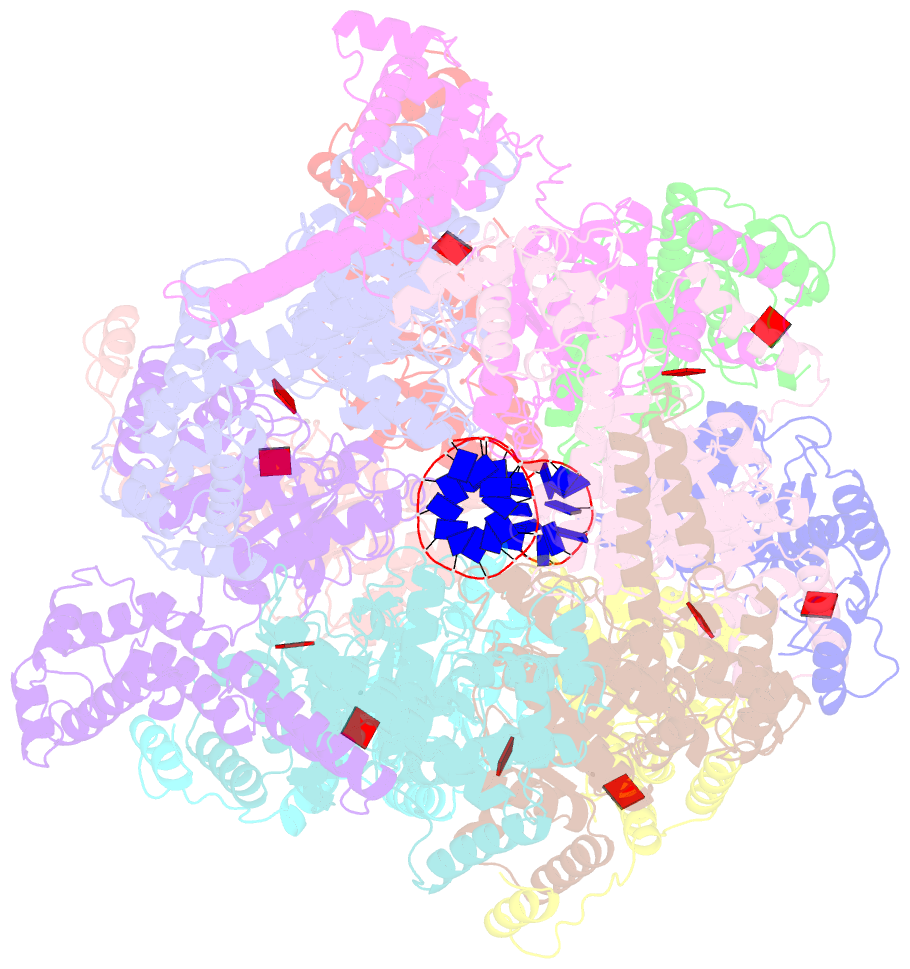
- E. coli dnabc complex bound to ssDNA
- Reference
- Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM (2019): "Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA." Mol.Cell, 74, 173-184.e4. doi: 10.1016/j.molcel.2019.01.023.
- Abstract
- In cells, dedicated AAA+ ATPases deposit hexameric, ring-shaped helicases onto DNA to initiate chromosomal replication. To better understand the mechanisms by which helicase loading can occur, we used cryo-EM to determine sub-4-Å-resolution structures of the E. coli DnaB⋅DnaC helicase⋅loader complex with nucleotide in pre- and post-DNA engagement states. In the absence of DNA, six DnaC protomers latch onto and crack open a DnaB hexamer using an extended N-terminal domain, stabilizing this conformation through nucleotide-dependent ATPase interactions. Upon binding DNA, DnaC hydrolyzes ATP, allowing DnaB to isomerize into a topologically closed, pre-translocation state competent to bind primase. Our data show how DnaC opens the DnaB ring and represses the helicase prior to DNA binding and how DnaC ATPase activity is reciprocally regulated by DnaB and DNA. Comparative analyses reveal how the helicase loading mechanism of DnaC parallels and diverges from homologous AAA+ systems involved in DNA replication and transposition.