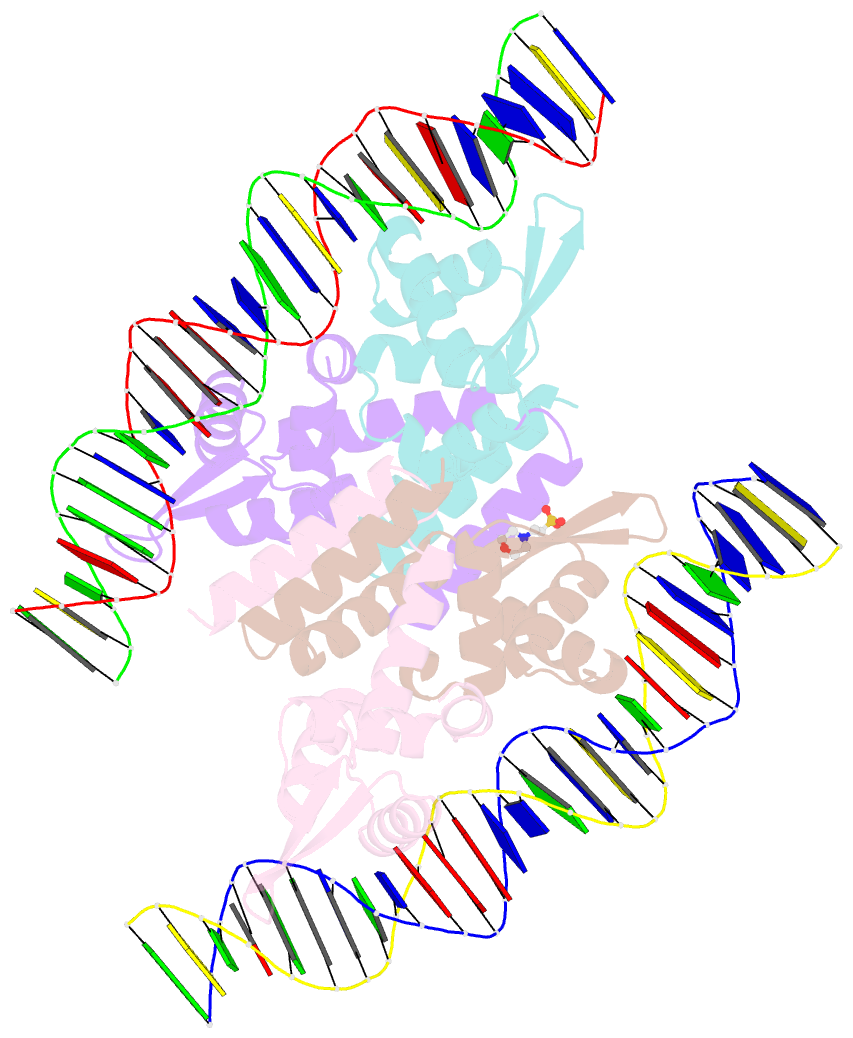
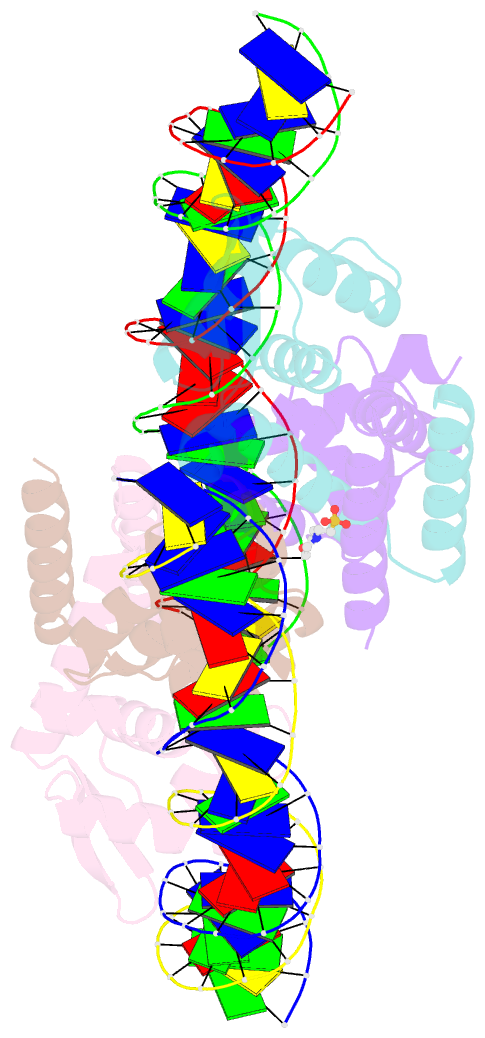
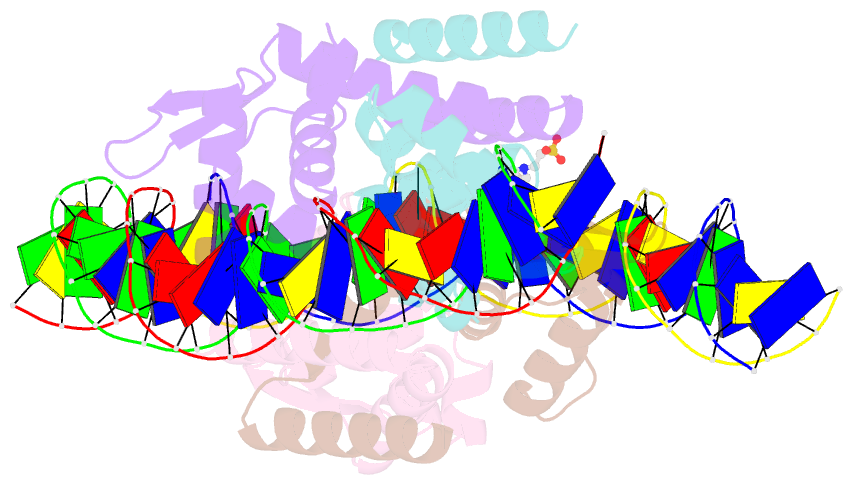
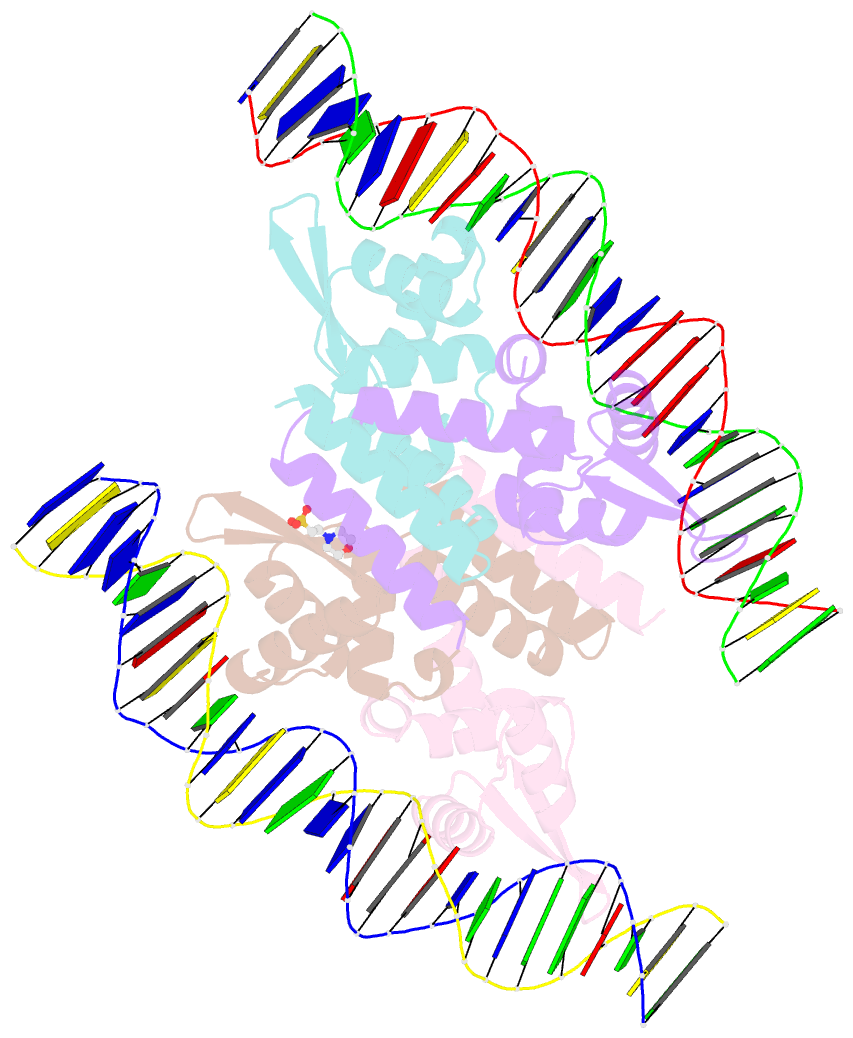
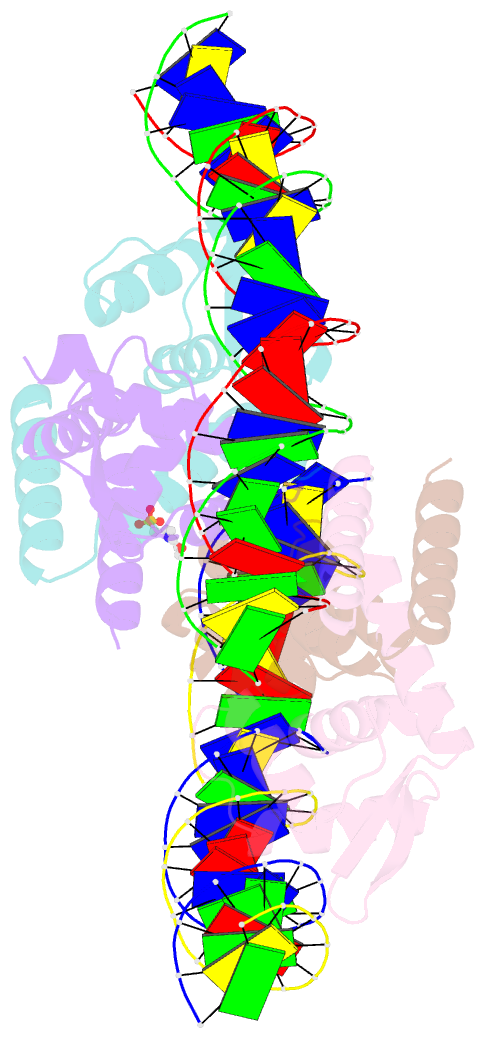
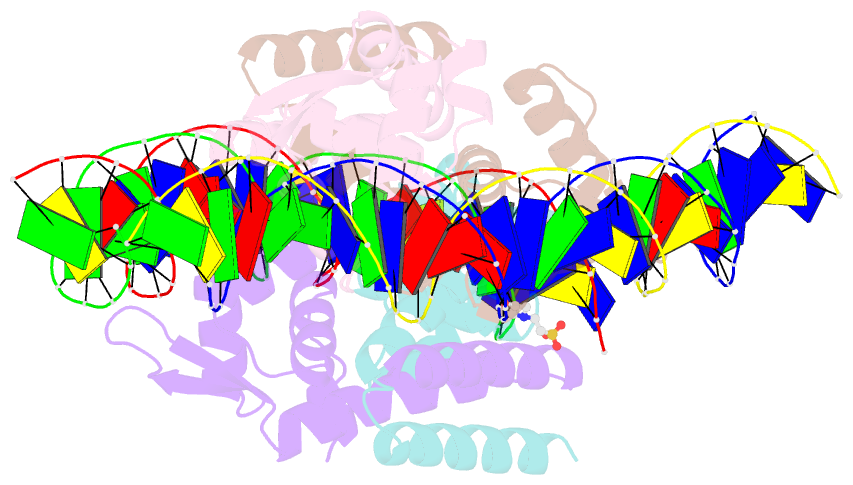
Summary information and primary citation
- PDB-id
- 6qil; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.0 Å)
- Summary
- The complex structure of hsrosr-s1 (vng0258h-rosr-s1)
- Reference
- Kutnowski N, Shmulevich F, Davidov G, Shahar A, Bar-Zvi D, Eichler J, Zarivach R, Shaanan B (2019): "Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR." Nucleic Acids Res., 47, 8860-8873. doi: 10.1093/nar/gkz604.
- Abstract
- Interactions between proteins and DNA are crucial for all biological systems. Many studies have shown the dependence of protein-DNA interactions on the surrounding salt concentration. How these interactions are maintained in the hypersaline environments that halophiles inhabit remains puzzling. Towards solving this enigma, we identified the DNA motif recognized by the Halobactrium salinarum ROS-dependent transcription factor (hsRosR), determined the structure of several hsRosR-DNA complexes and investigated the DNA-binding process under extreme high-salt conditions. The picture that emerges from this work contributes to our understanding of the principles underlying the interplay between electrostatic interactions and salt-mediated protein-DNA interactions in an ionic environment characterized by molar salt concentrations.