Summary information and primary citation
- PDB-id
- 6r5k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- cryo-EM (4.8 Å)
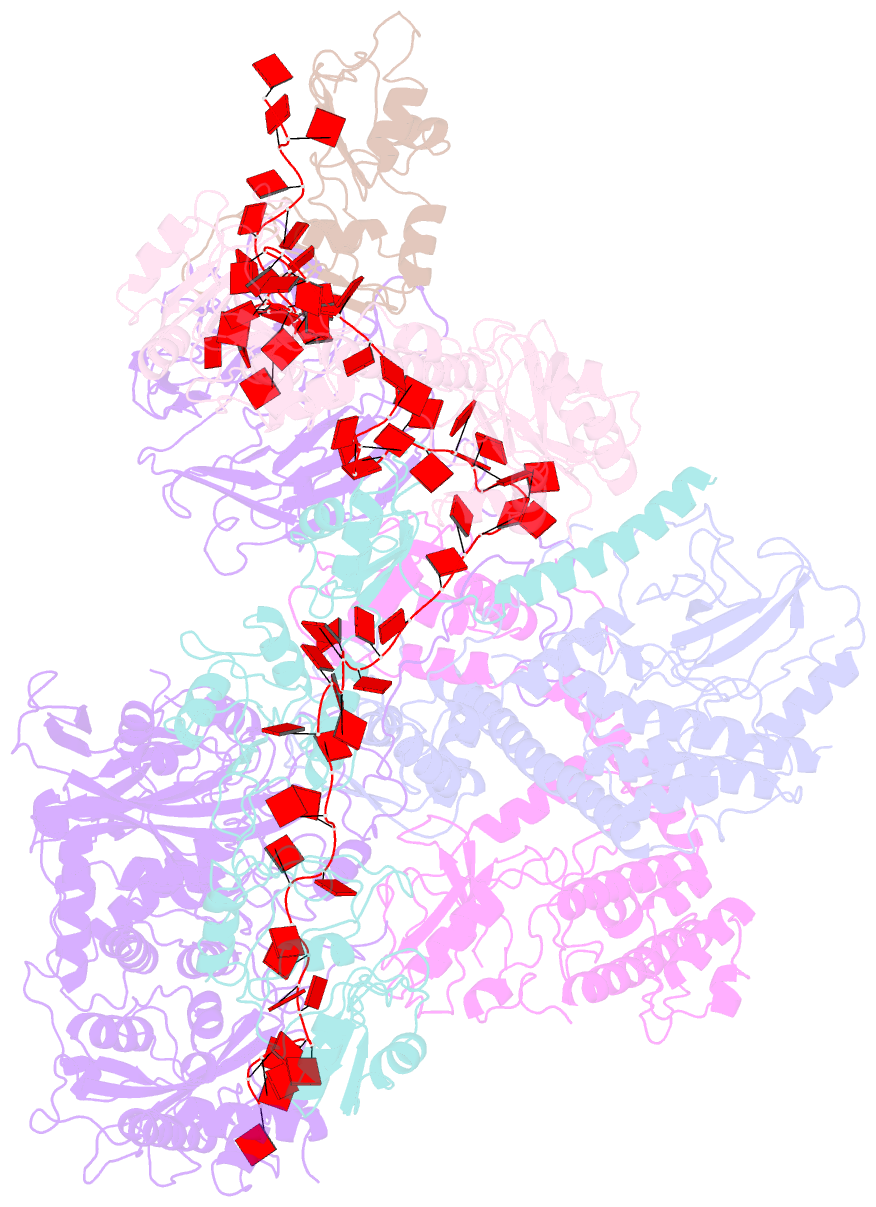
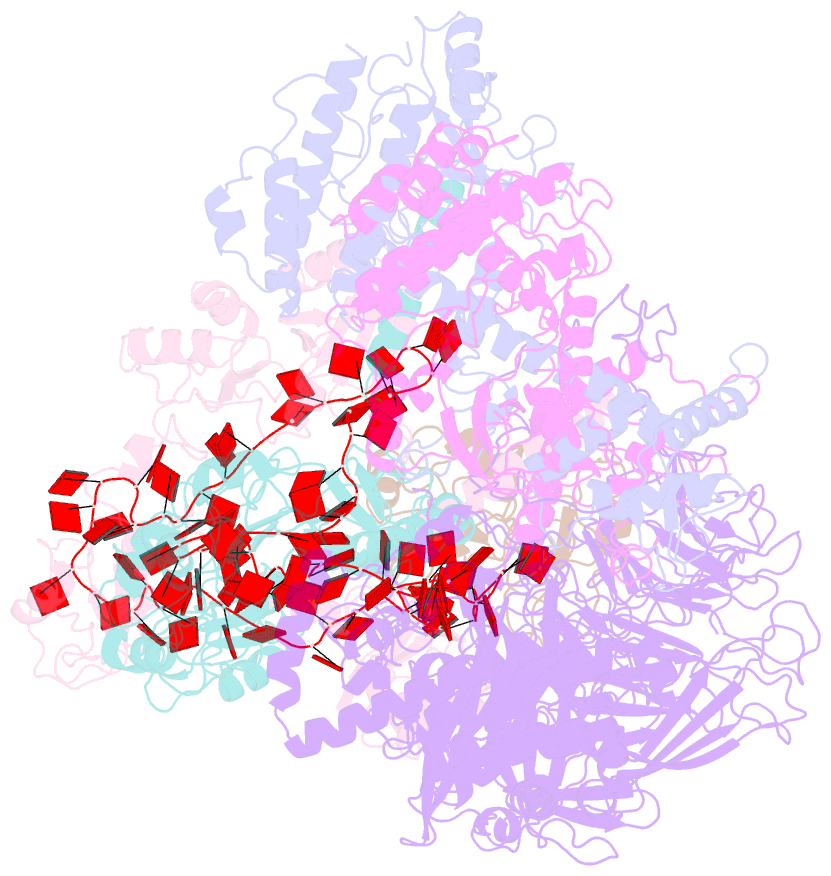
- Summary
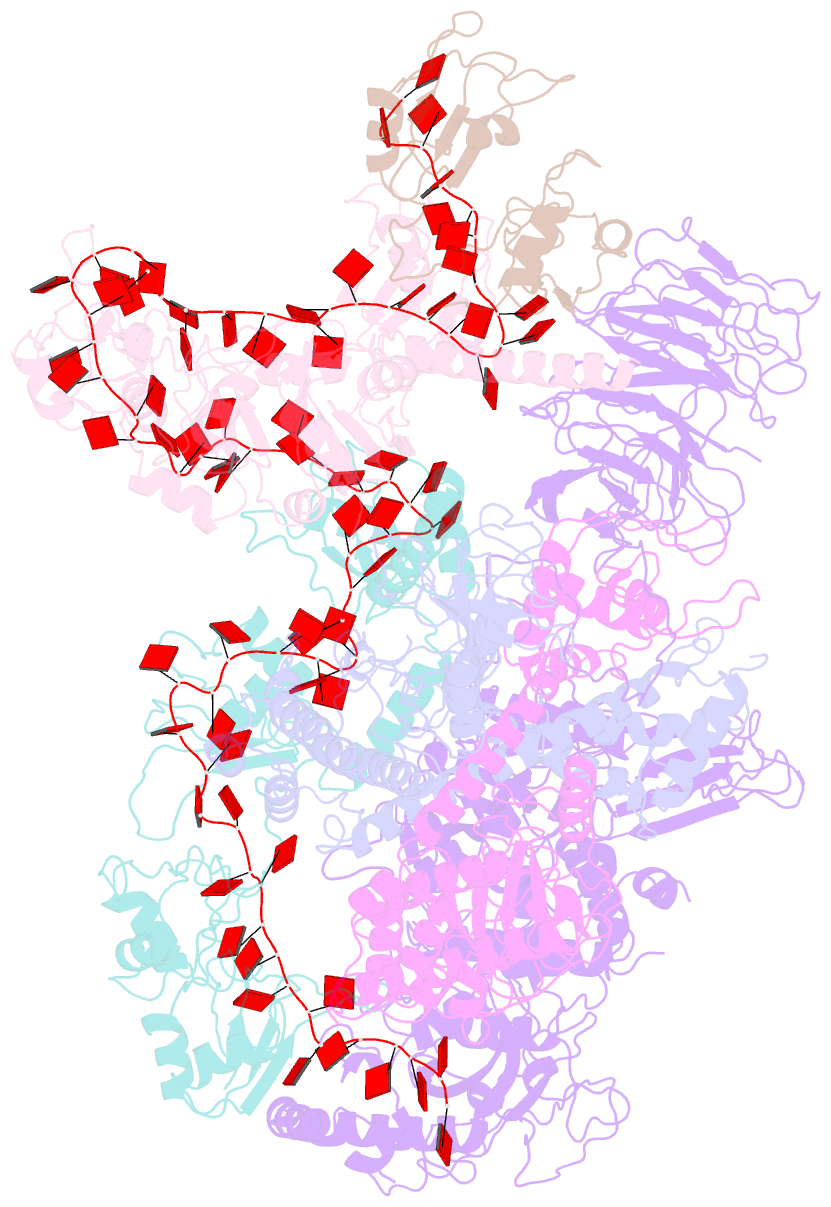
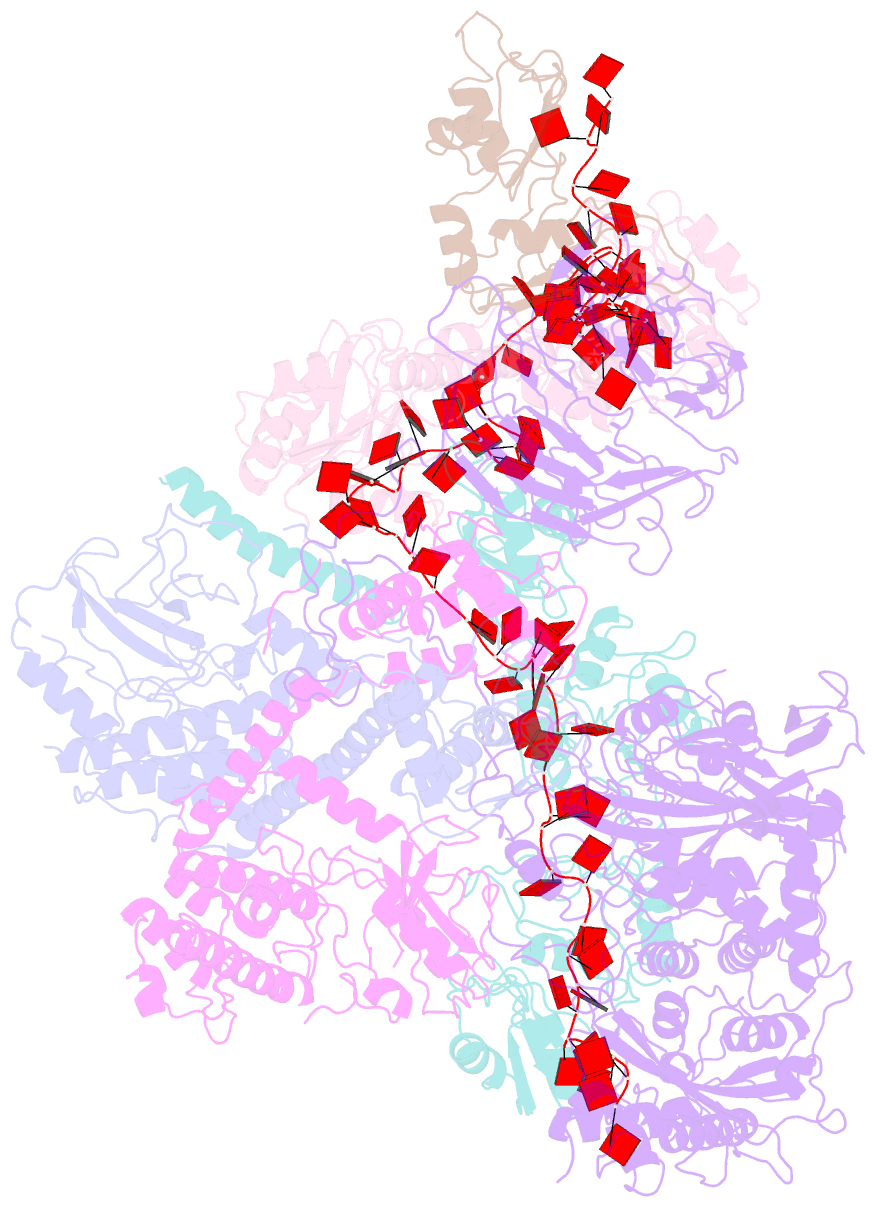
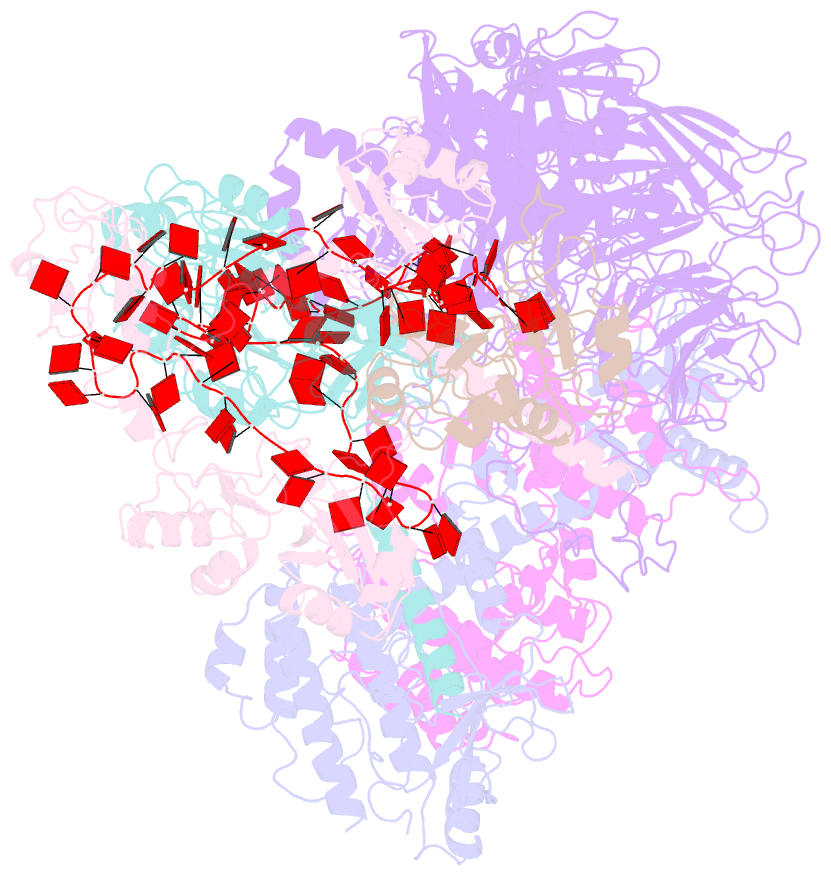
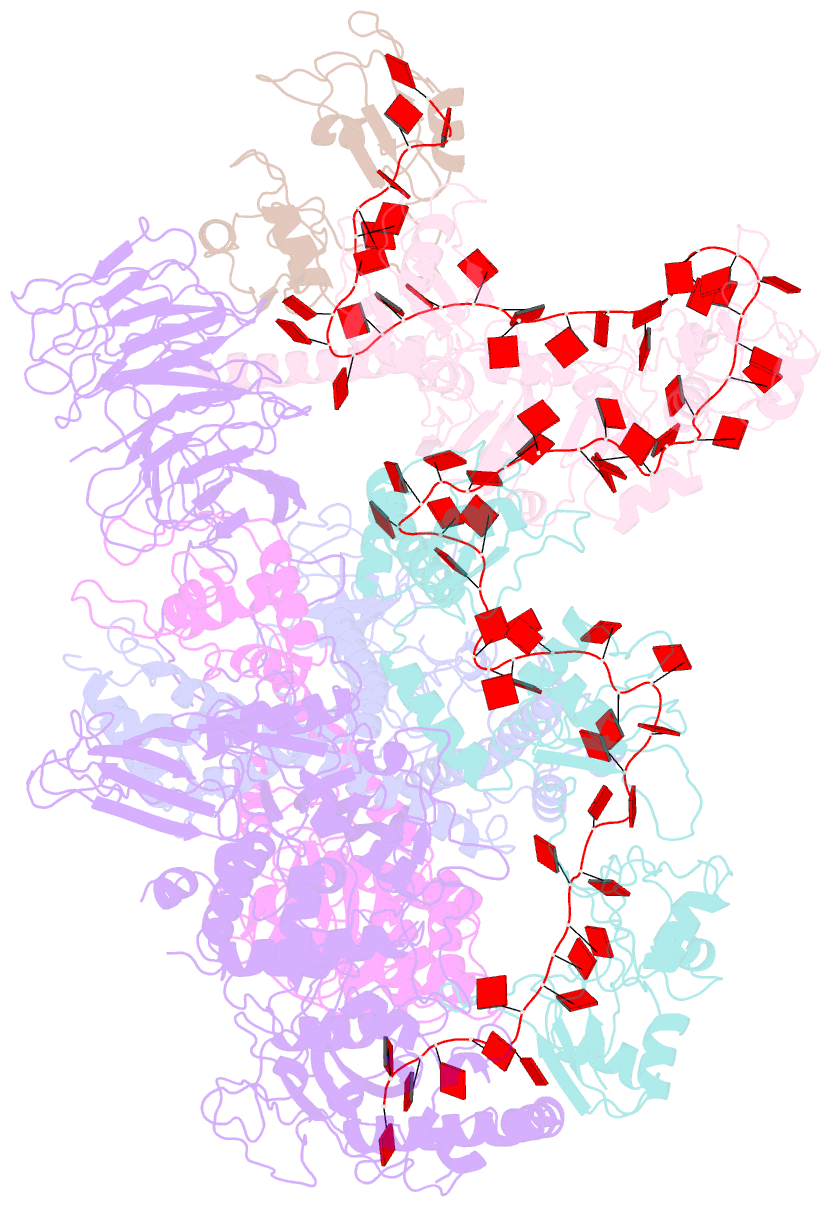
- cryo-EM structure of a poly(a) rnp bound to the pan2-pan3 deadenylase
- Reference
- Schafer IB, Yamashita M, Schuller JM, Schussler S, Reichelt P, Strauss M, Conti E (2019): "Molecular Basis for poly(A) RNP Architecture and Recognition by the Pan2-Pan3 Deadenylase." Cell, 177, 1619. doi: 10.1016/j.cell.2019.04.013.
- Abstract
- The stability of eukaryotic mRNAs is dependent on a ribonucleoprotein (RNP) complex of poly(A)-binding proteins (PABPC1/Pab1) organized on the poly(A) tail. This poly(A) RNP not only protects mRNAs from premature degradation but also stimulates the Pan2-Pan3 deadenylase complex to catalyze the first step of poly(A) tail shortening. We reconstituted this process in vitro using recombinant proteins and show that Pan2-Pan3 associates with and degrades poly(A) RNPs containing two or more Pab1 molecules. The cryo-EM structure of Pan2-Pan3 in complex with a poly(A) RNP composed of 90 adenosines and three Pab1 protomers shows how the oligomerization interfaces of Pab1 are recognized by conserved features of the deadenylase and thread the poly(A) RNA substrate into the nuclease active site. The structure reveals the basis for the periodic repeating architecture at the 3' end of cytoplasmic mRNAs. This illustrates mechanistically how RNA-bound Pab1 oligomers act as rulers for poly(A) tail length over the mRNAs' lifetime.