Summary information and primary citation
- PDB-id
- 6r9q; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (3.079 Å)
- Summary
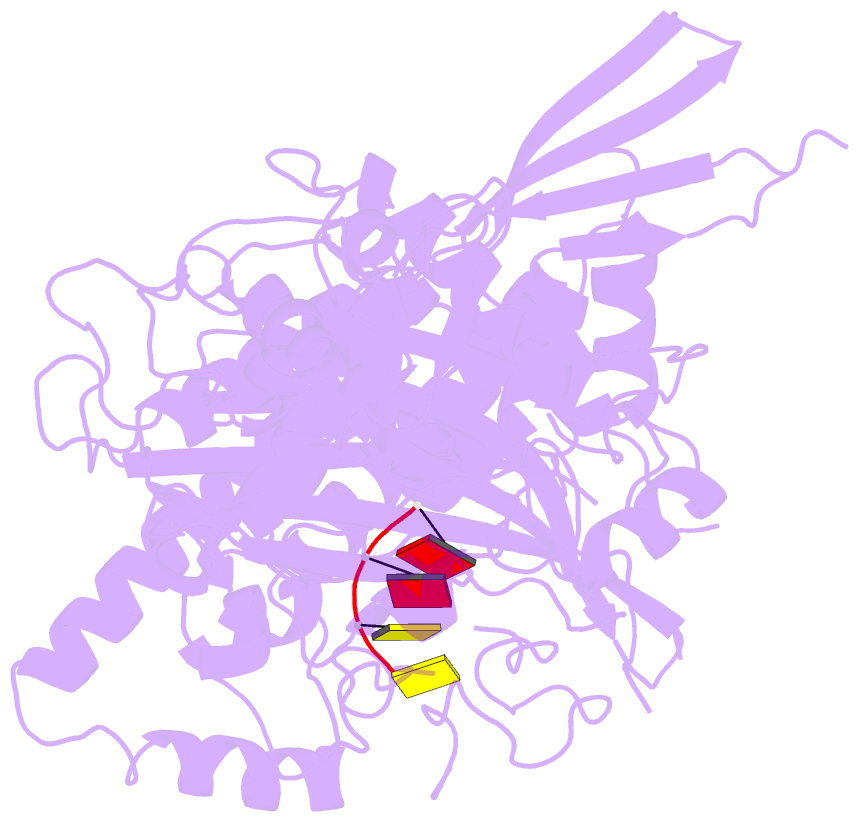
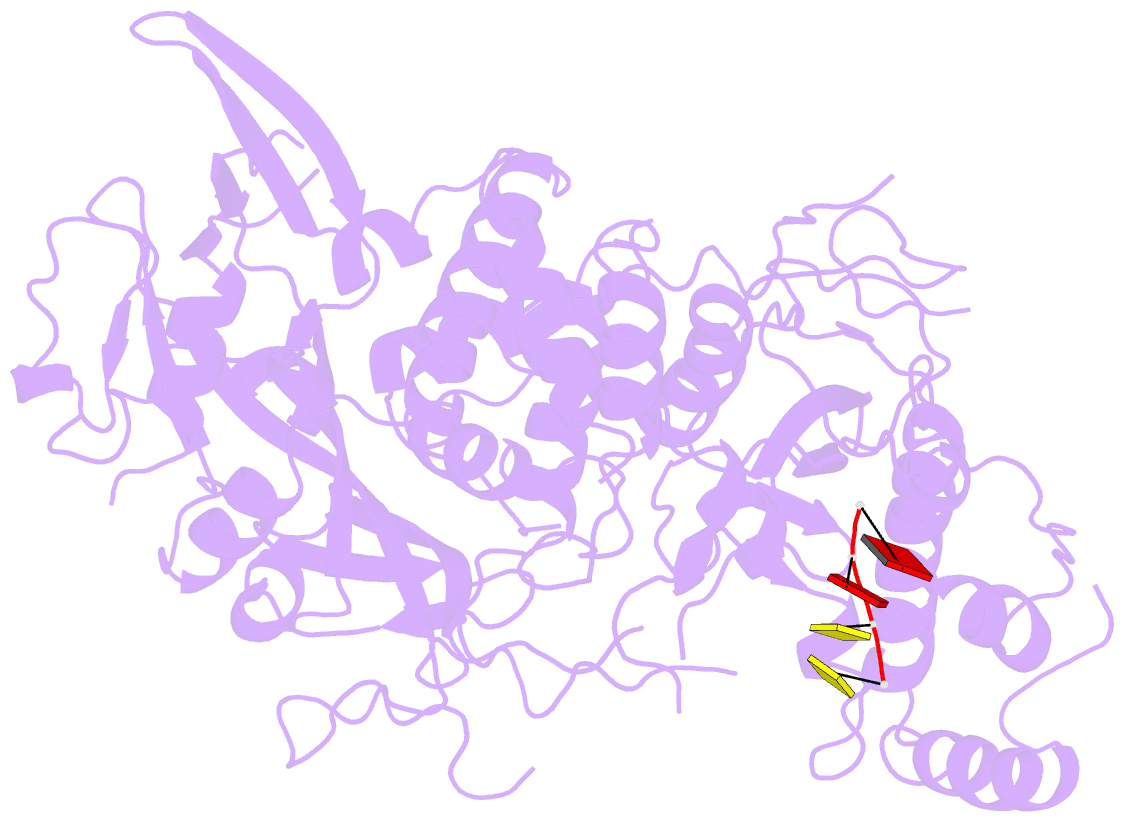
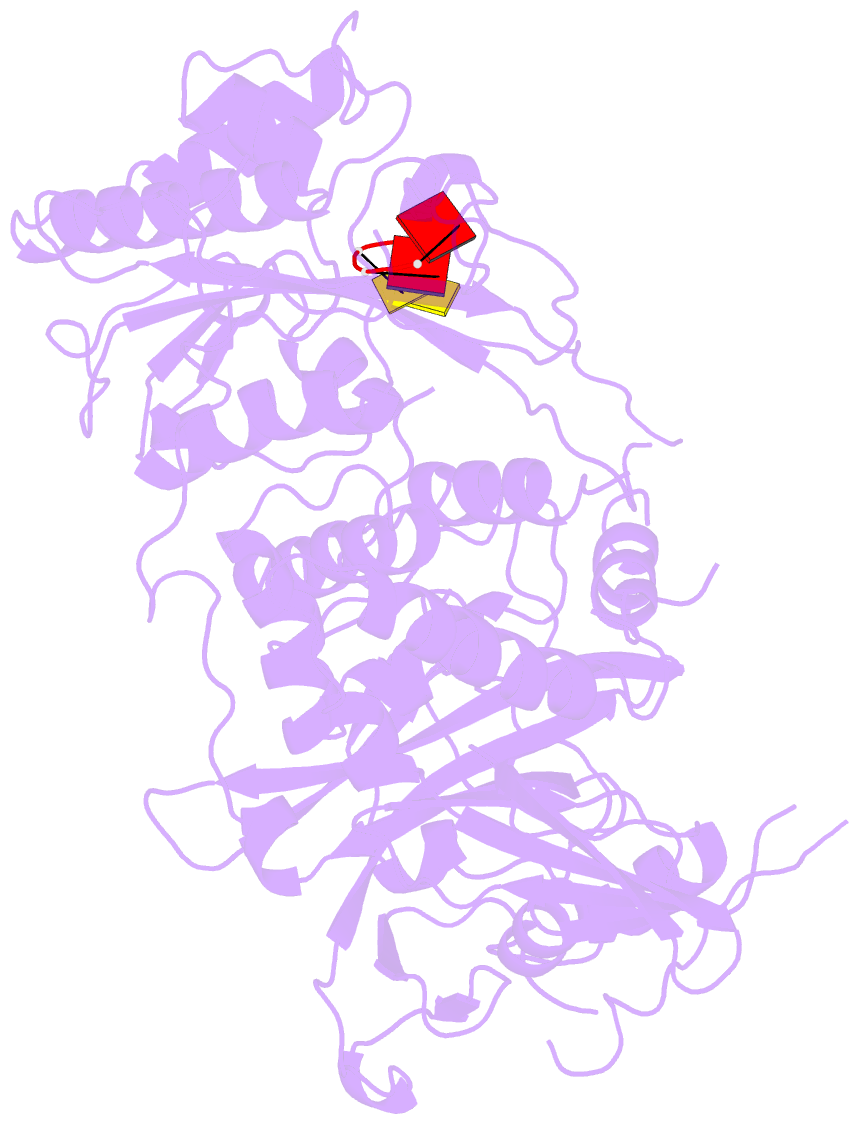
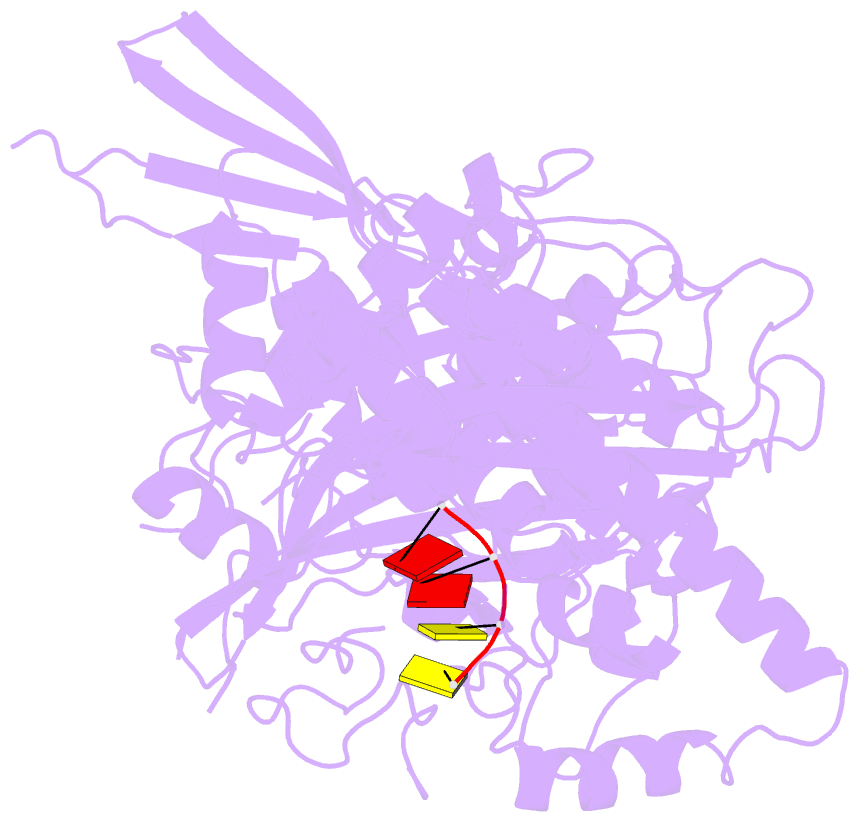
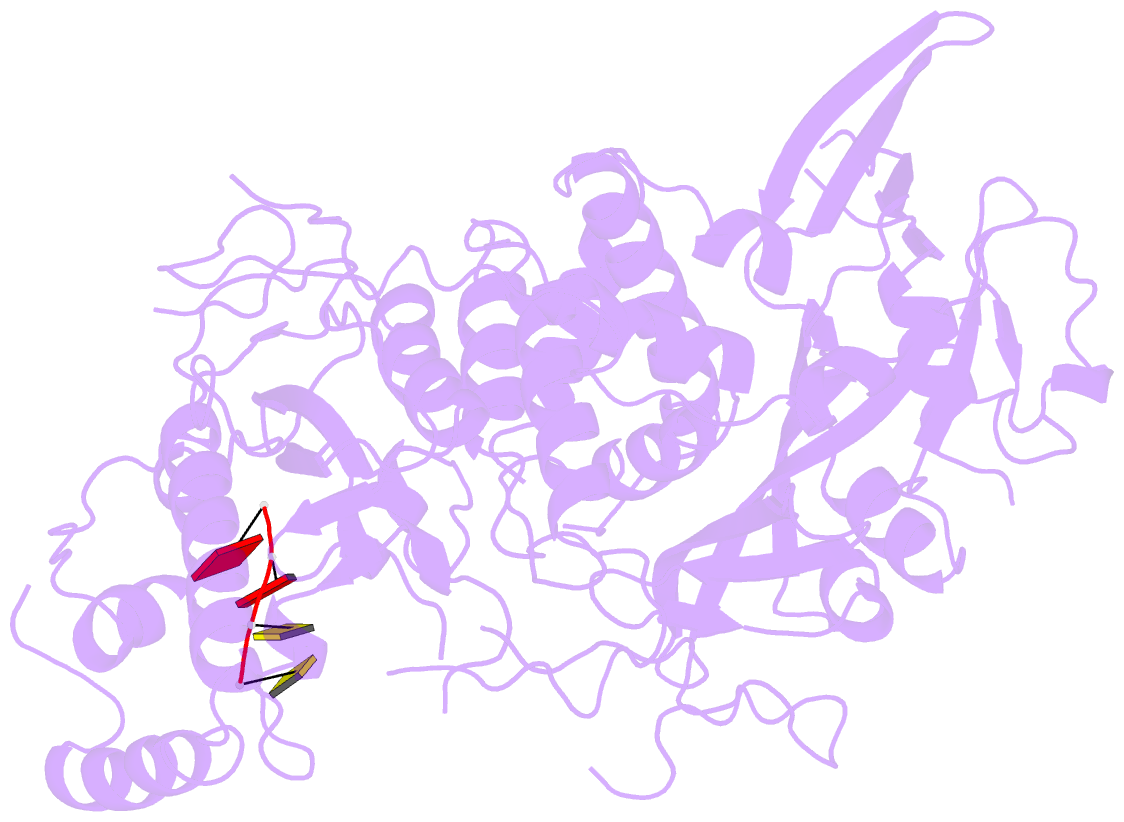
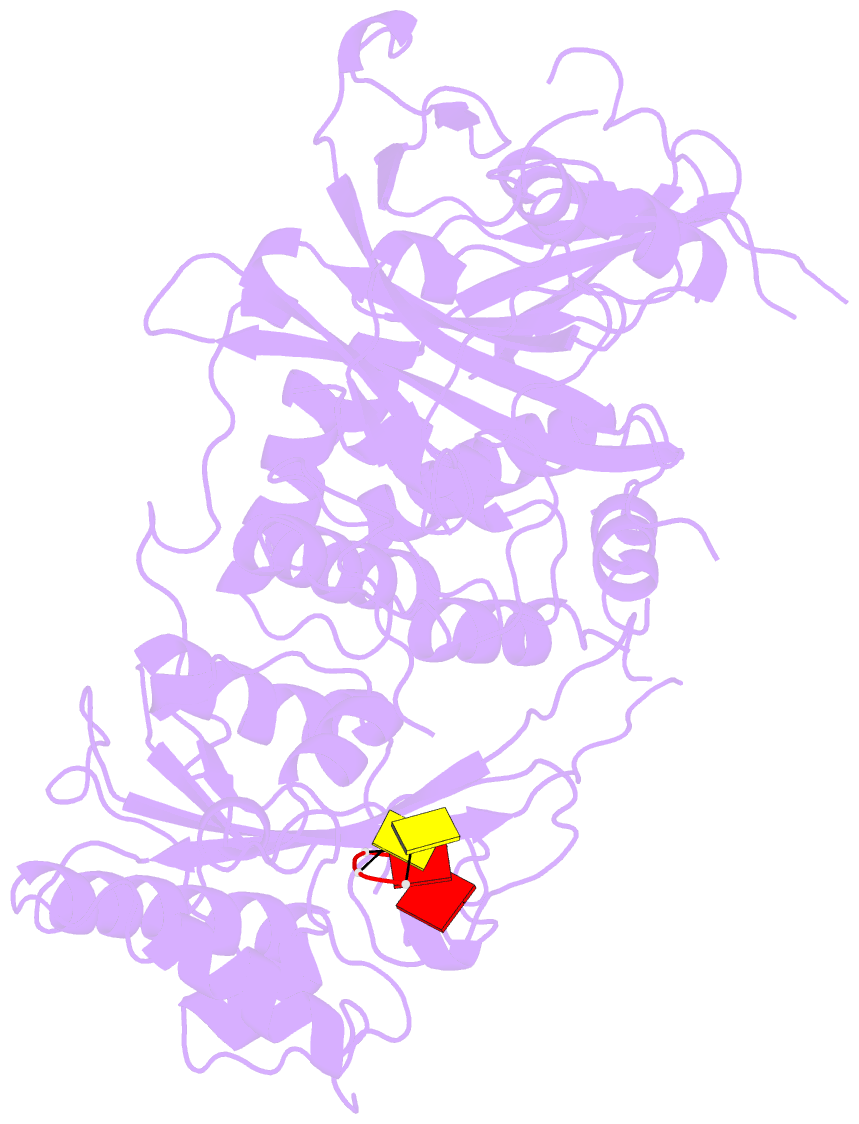
- Structure of saccharomyces cerevisiae apo pan2 pseudoubiquitin hydrolase-RNA exonuclease (uch-exo) module in complex with aaccaa RNA
- Reference
- Tang TTL, Stowell JAW, Hill CH, Passmore LA (2019): "The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases." Nat.Struct.Mol.Biol., 26, 433-442. doi: 10.1038/s41594-019-0227-9.
- Abstract
- The 3' poly(A) tail of messenger RNA is fundamental to regulating eukaryotic gene expression. Shortening of the poly(A) tail, termed deadenylation, reduces transcript stability and inhibits translation. Nonetheless, the mechanism for poly(A) recognition by the conserved deadenylase complexes Pan2-Pan3 and Ccr4-Not is poorly understood. Here we provide a model for poly(A) RNA recognition by two DEDD-family deadenylase enzymes, Pan2 and the Ccr4-Not nuclease Caf1. Crystal structures of Saccharomyces cerevisiae Pan2 in complex with RNA show that, surprisingly, Pan2 does not form canonical base-specific contacts. Instead, it recognizes the intrinsic stacked, helical conformation of poly(A) RNA. Using a fully reconstituted biochemical system, we show that disruption of this structure-for example, by incorporation of guanosine into poly(A)-inhibits deadenylation by both Pan2 and Caf1. Together, these data establish a paradigm for specific recognition of the conformation of poly(A) RNA by proteins that regulate gene expression.