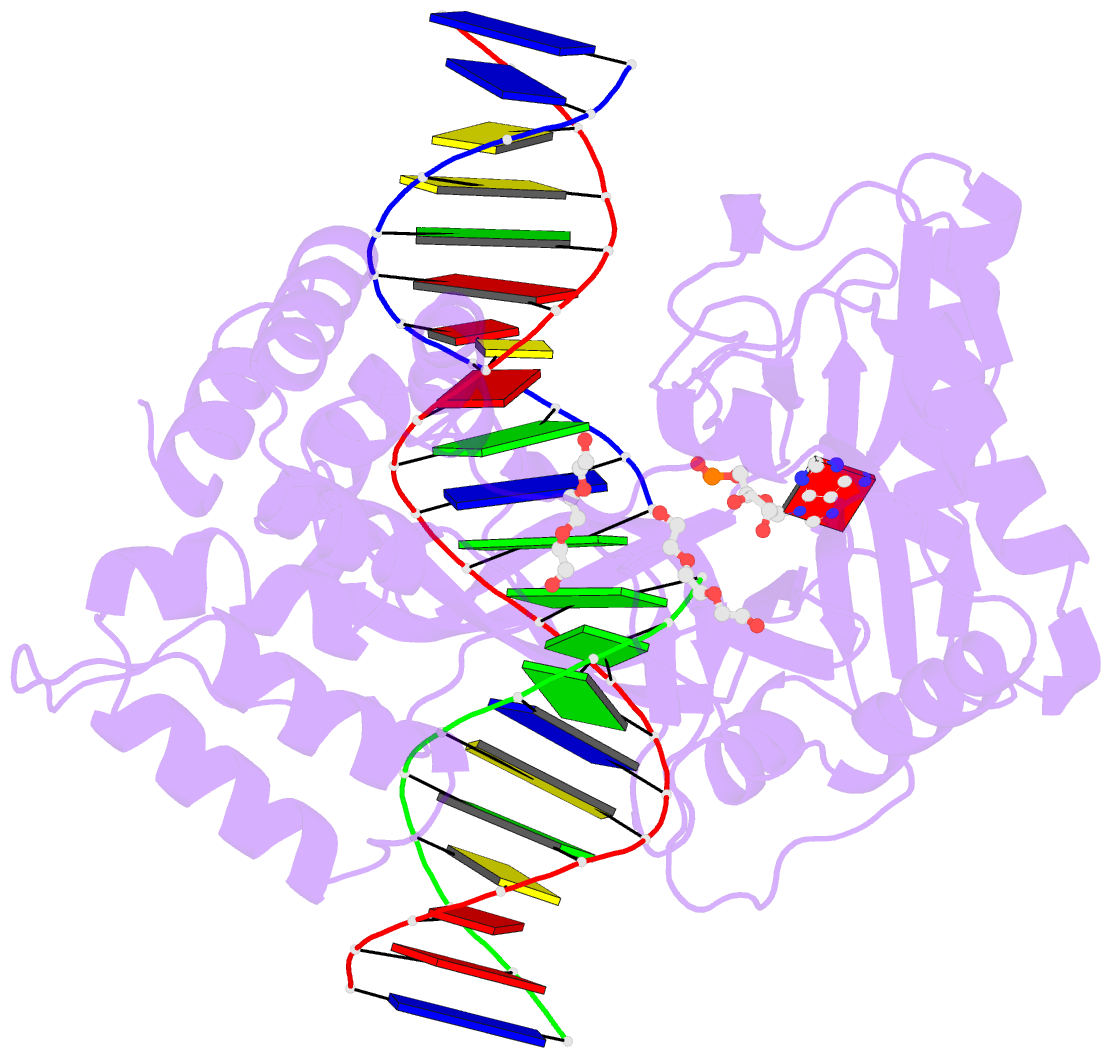
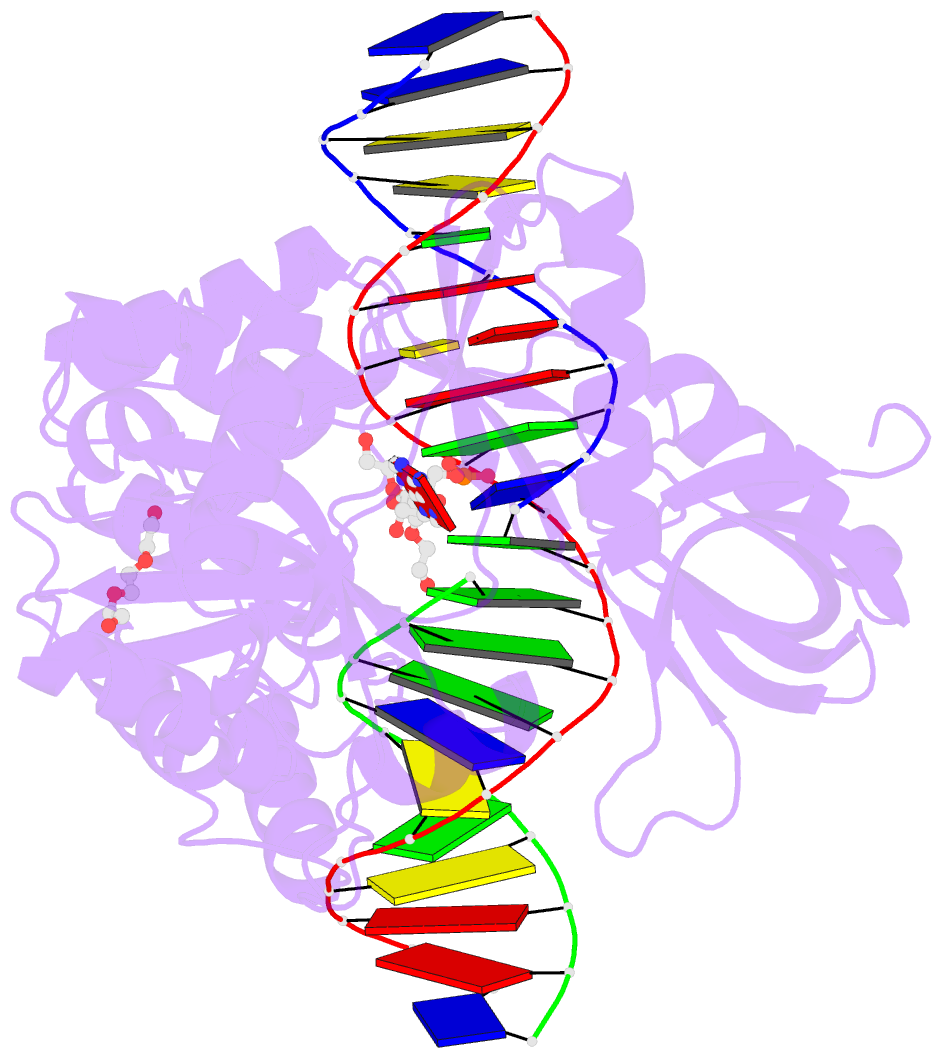
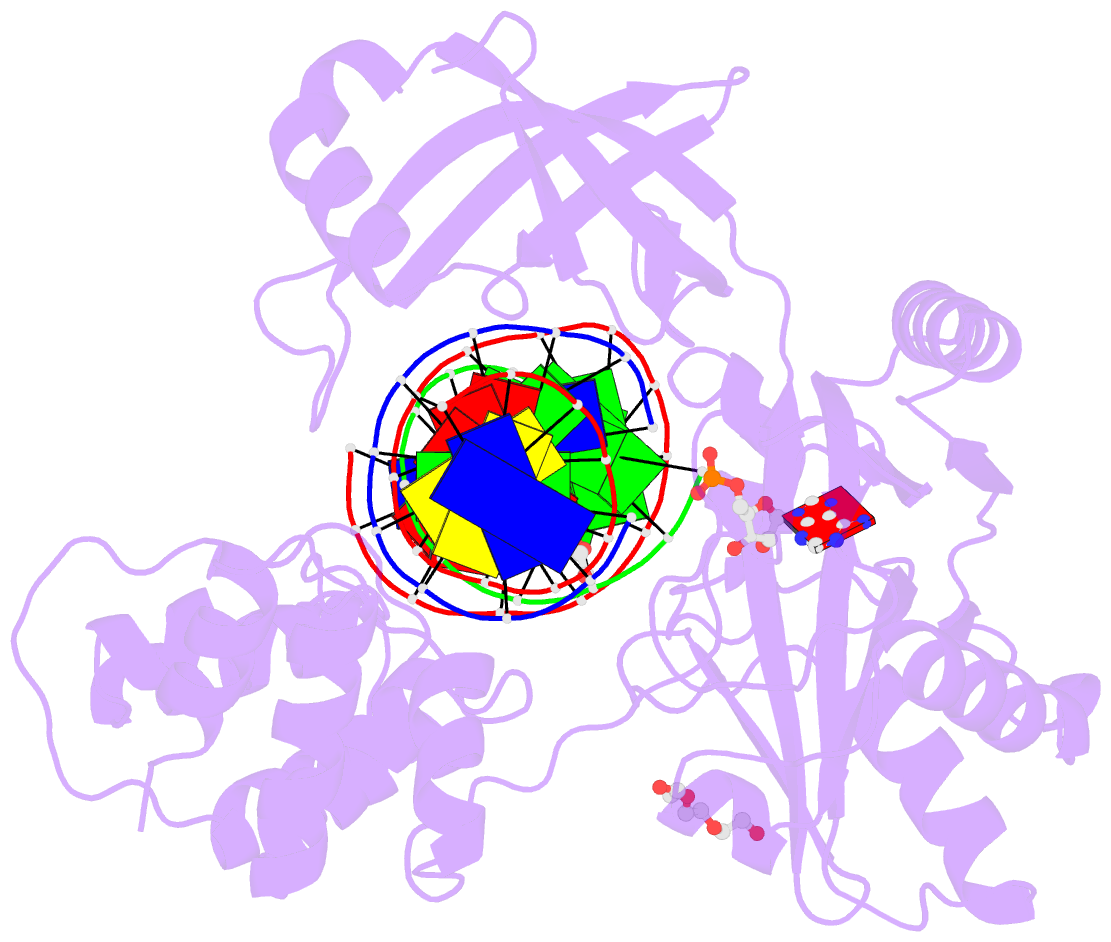
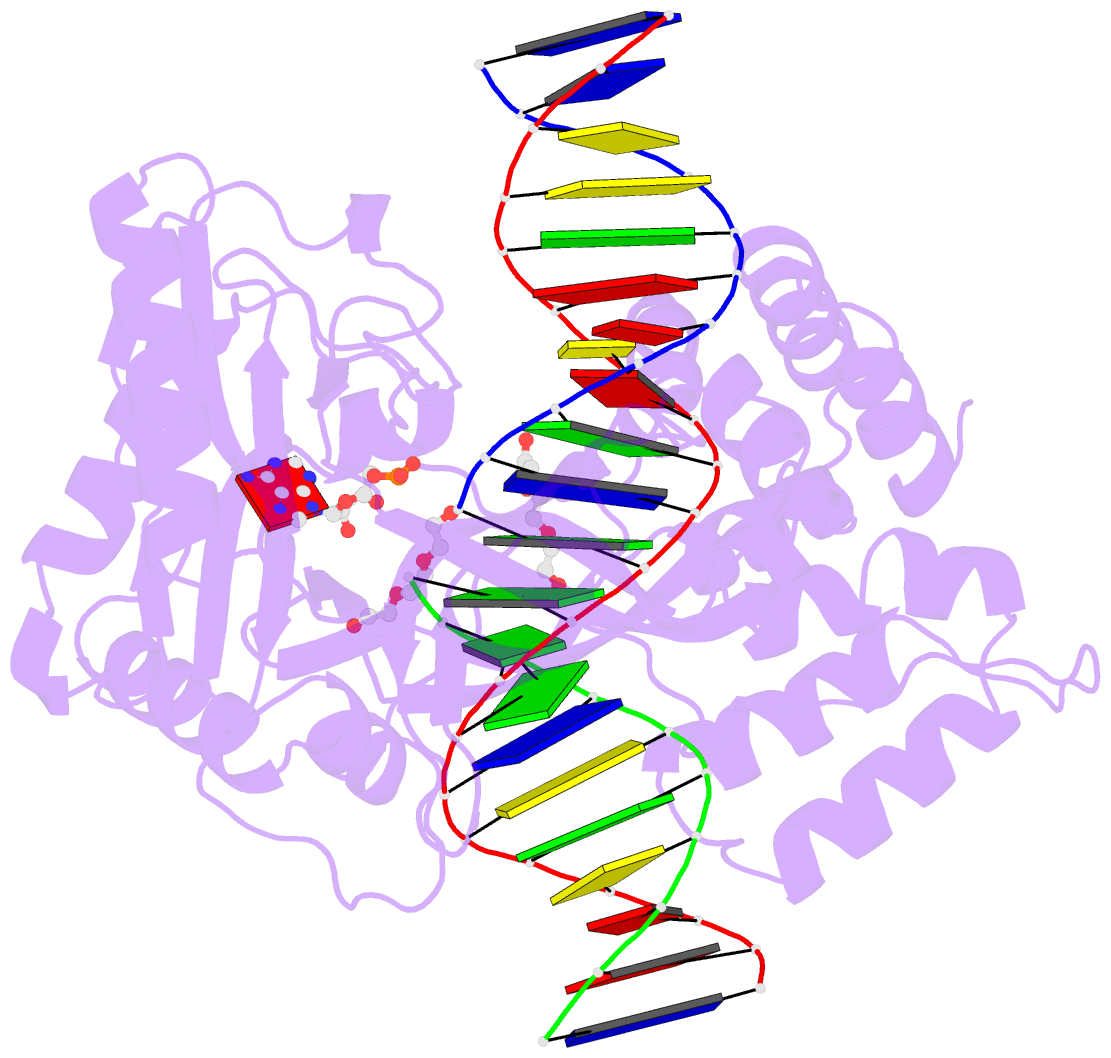
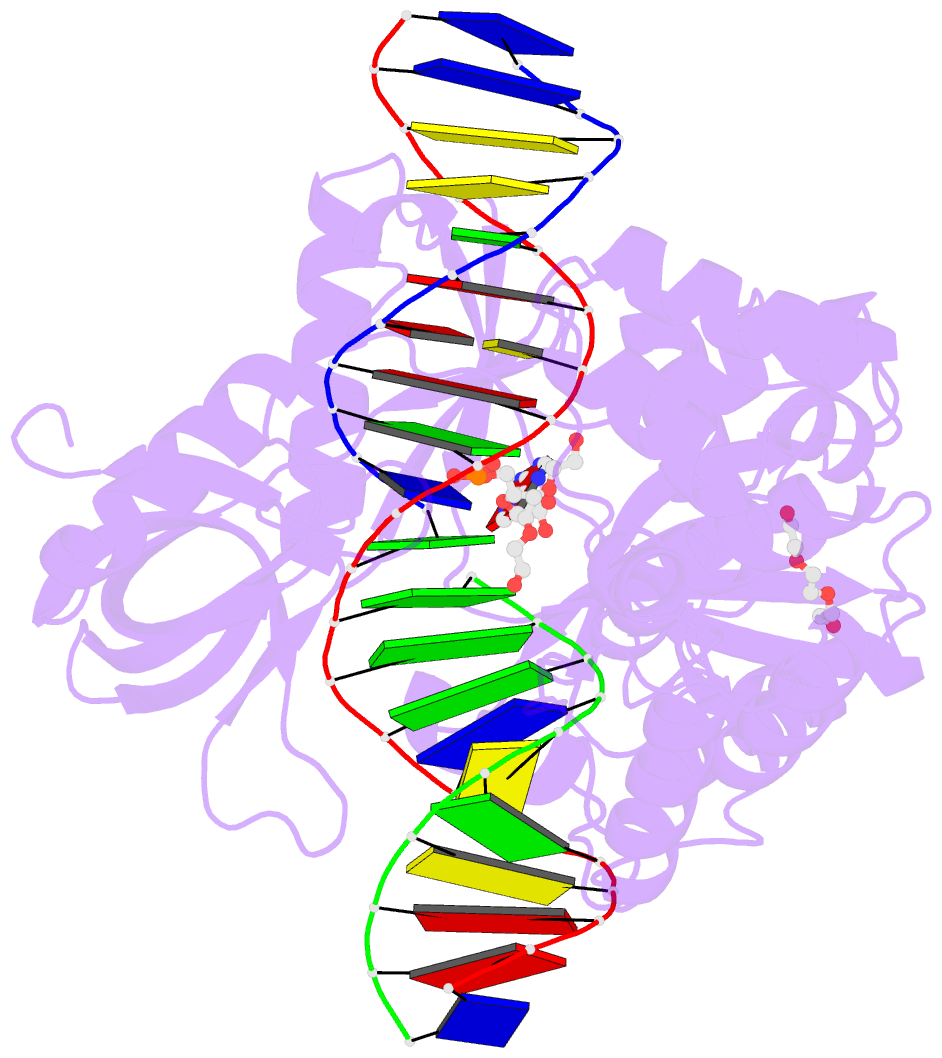
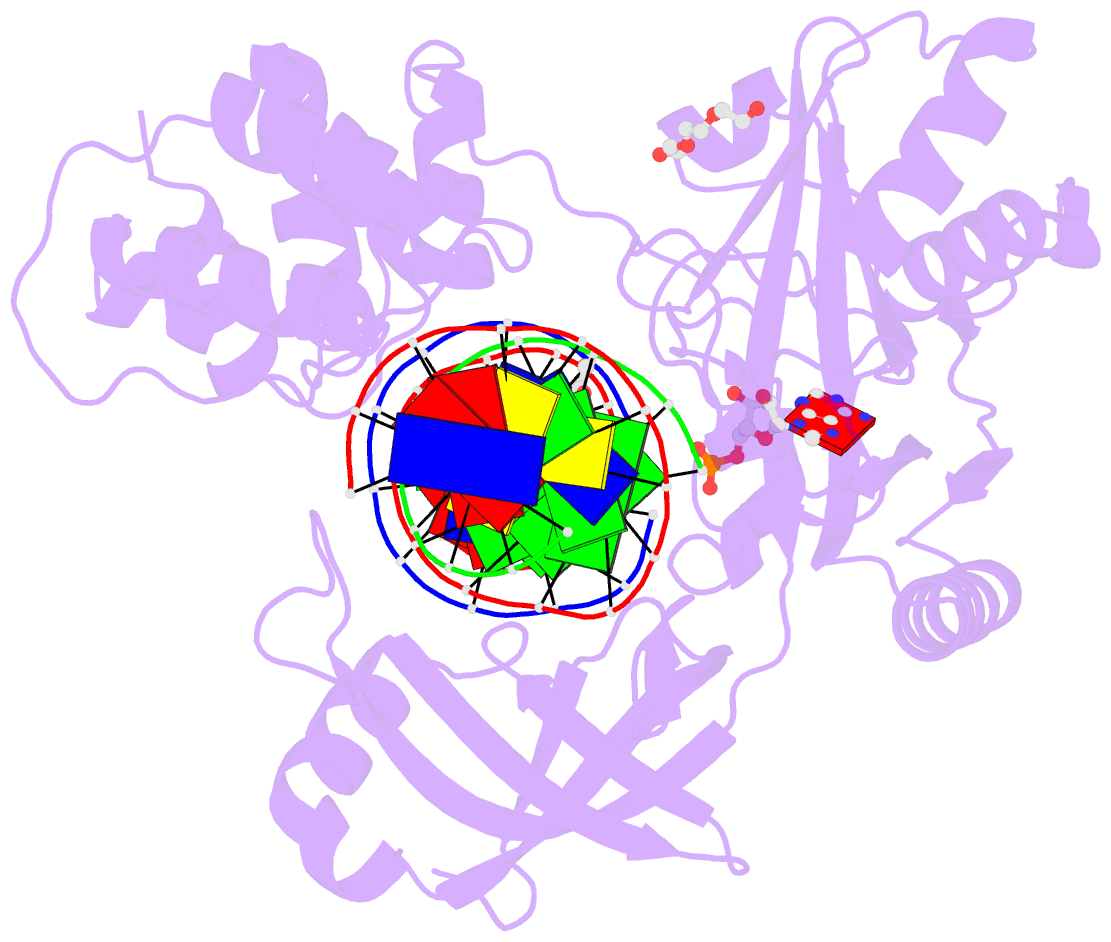
Summary information and primary citation
- PDB-id
- 6ras; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.75 Å)
- Summary
- Pmar-lig_pre.
- Reference
- Williamson A, Leiros HS (2019): "Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation." Nucleic Acids Res., 47, 7147-7162. doi: 10.1093/nar/gkz596.
- Abstract
- DNA ligases join adjacent 5' phosphate (5'P) and 3' hydroxyl (3'OH) termini of double-stranded DNA via a three-step mechanism requiring a nucleotide cofactor and divalent metal ion. Although considerable structural detail is available for the first two steps, less is known about step 3 where the DNA-backbone is joined or about the cation role at this step. We have captured high-resolution structures of an adenosine triphosphate (ATP)-dependent DNA ligase from Prochlorococcus marinus including a Mn-bound pre-ternary ligase-DNA complex poised for phosphodiester bond formation, and a post-ternary intermediate retaining product DNA and partially occupied AMP in the active site. The pre-ternary structure unambiguously identifies the binding site of the catalytic metal ion and confirms both its role in activating the 3'OH terminus for nucleophilic attack on the 5'P group and stabilizing the pentavalent transition state. The post-ternary structure indicates that DNA distortion and most enzyme-AMP contacts remain after phosphodiester bond formation, implying loss of covalent linkage to the DNA drives release of AMP, rather than active site rearrangement. Additionally, comparisons of this cyanobacterial DNA ligase with homologs from bacteria and bacteriophage pose interesting questions about the structural origin of double-strand break joining activity and the evolution of these ATP-dependent DNA ligase enzymes.