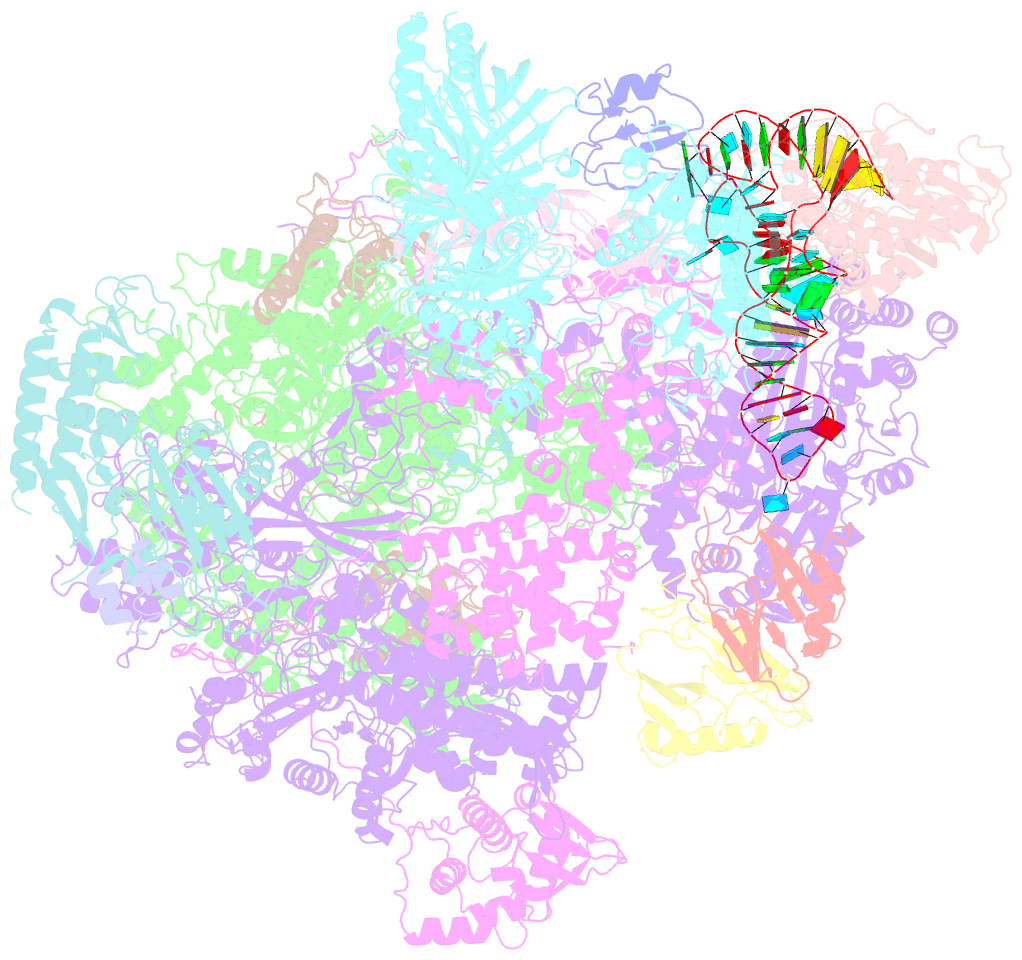
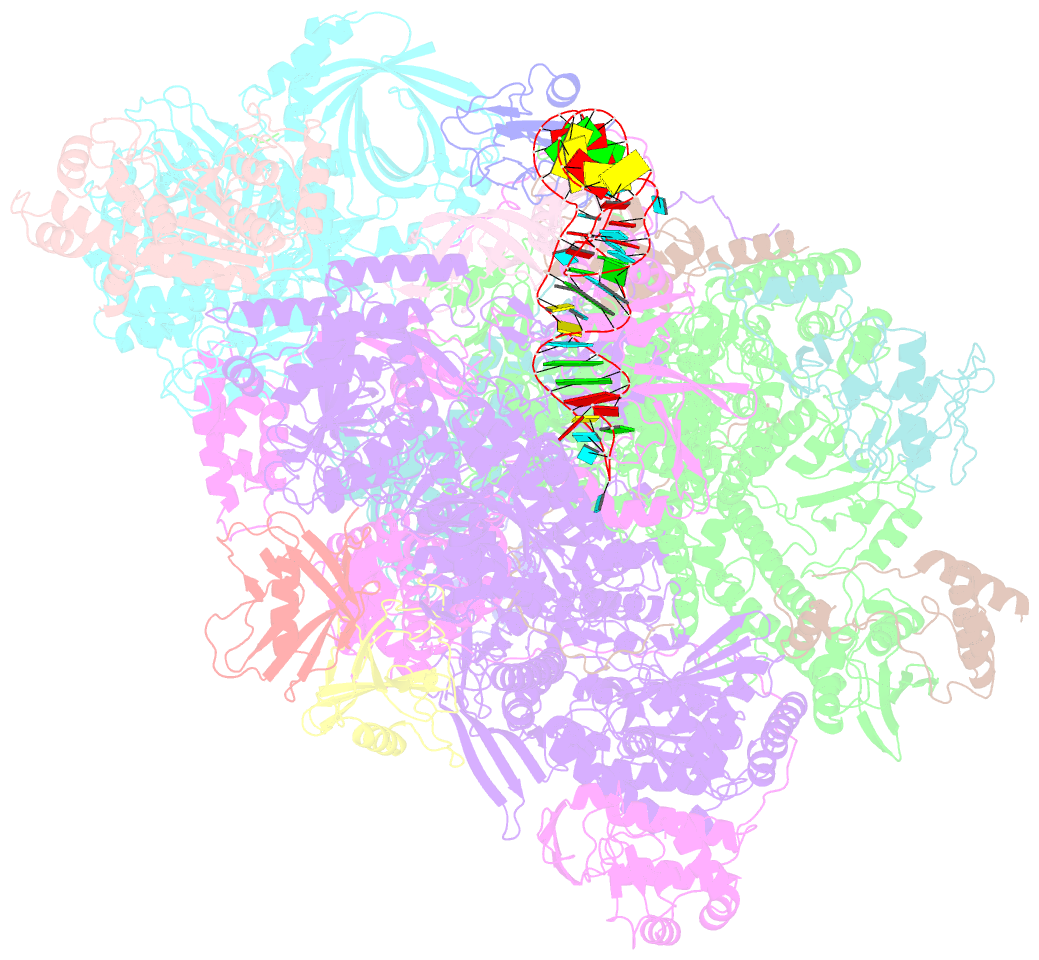
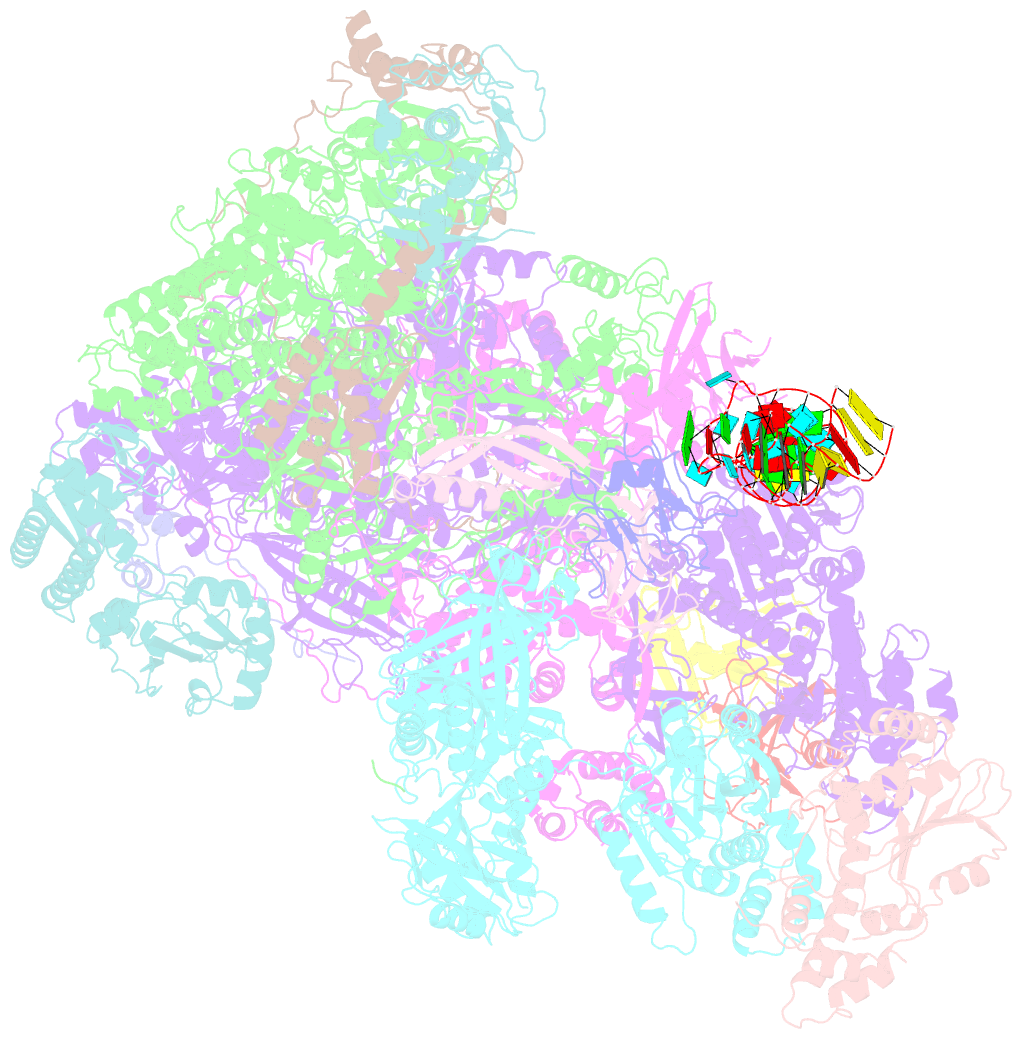
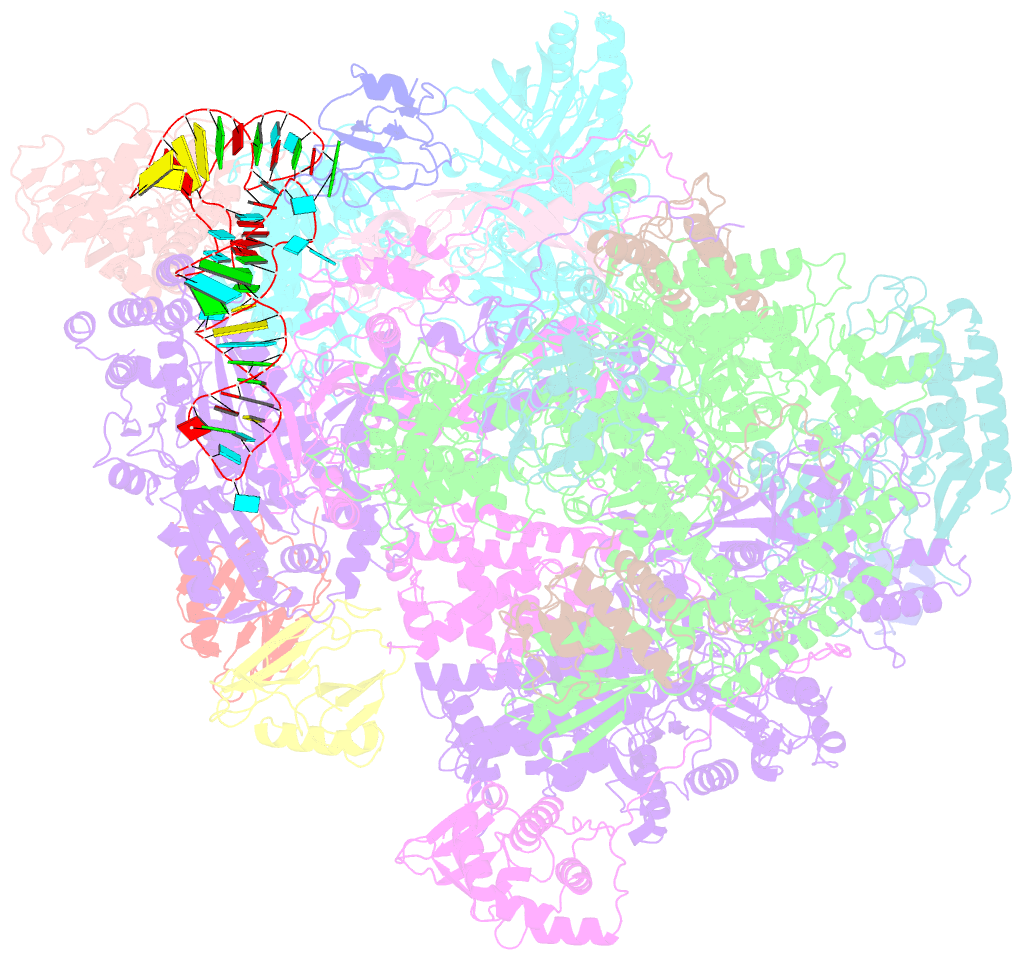
Summary information and primary citation
- PDB-id
- 6rfl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (2.76 Å)
- Summary
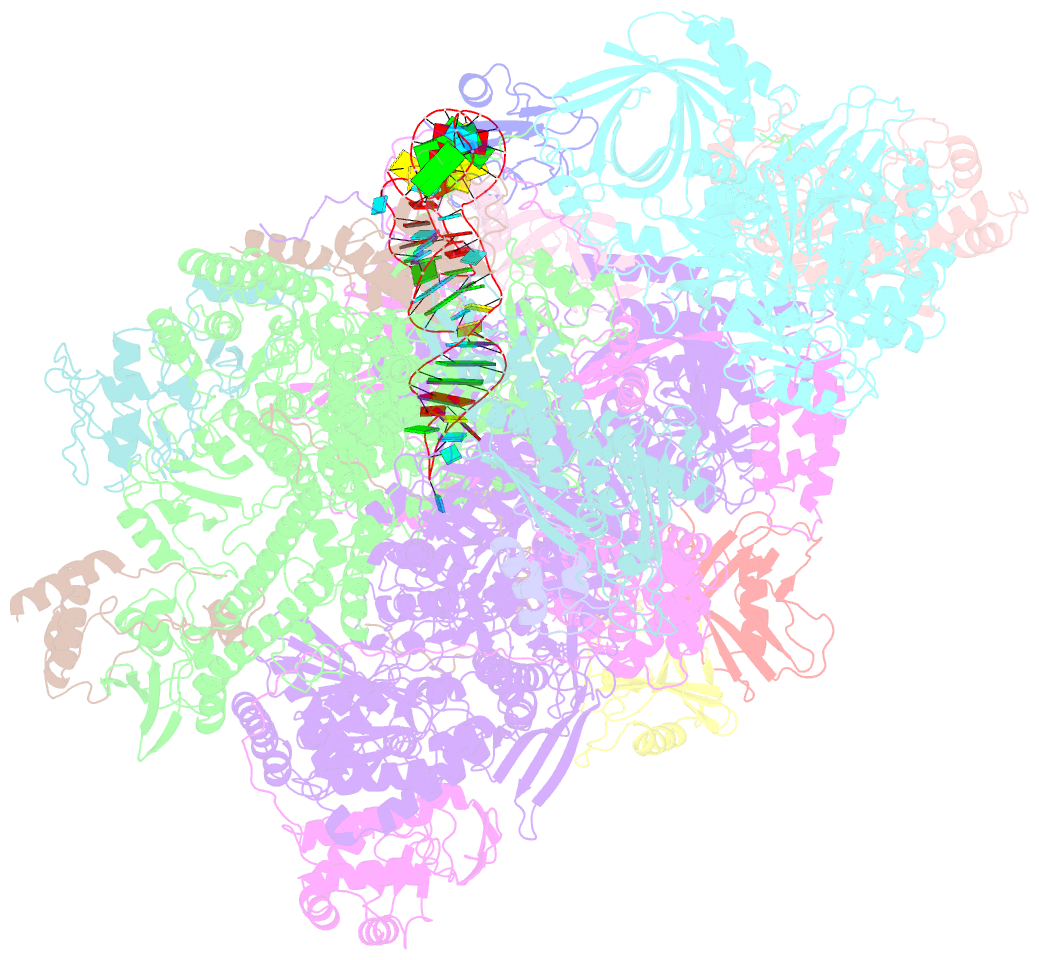
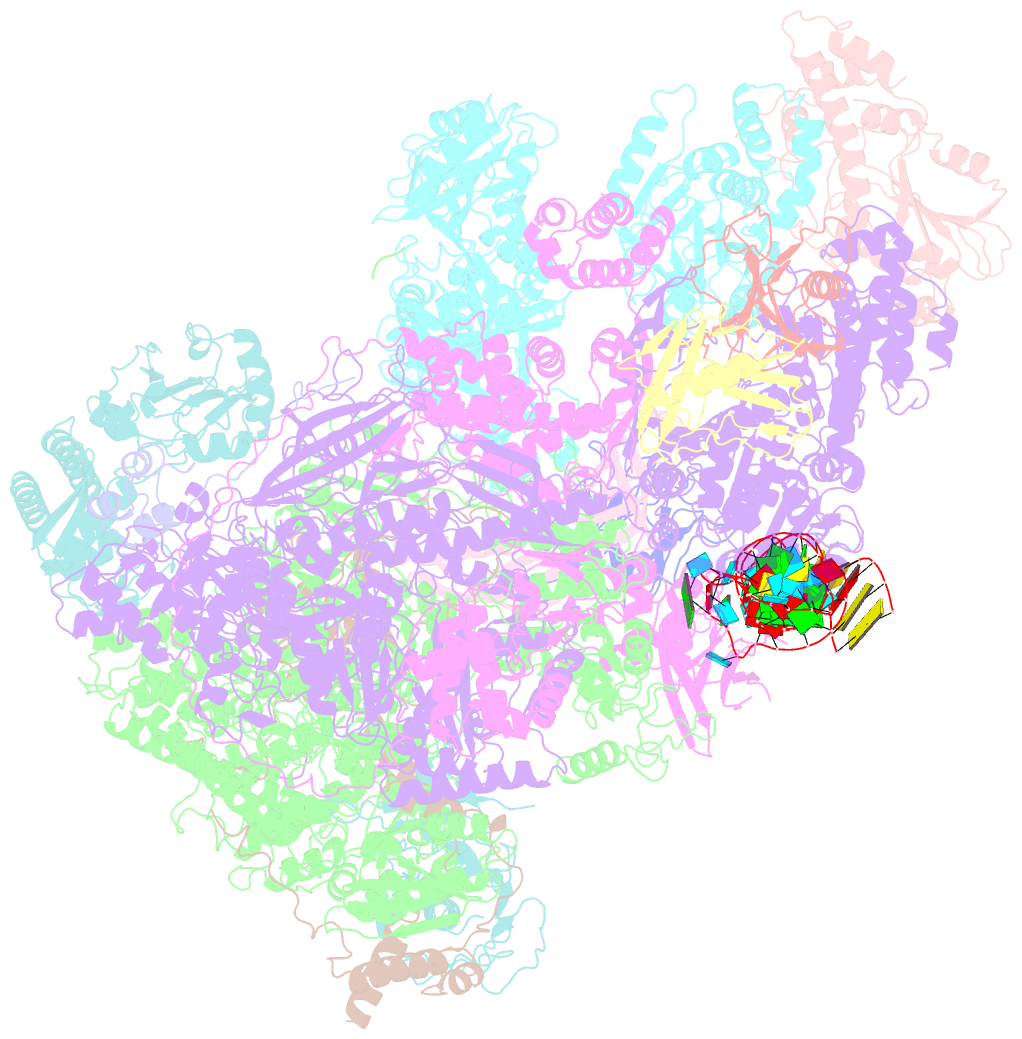
- Structure of the complete vaccinia DNA-dependent RNA polymerase complex
- Reference
- Grimm C, Hillen HS, Bedenk K, Bartuli J, Neyer S, Zhang Q, Huttenhofer A, Erlacher M, Dienemann C, Schlosser A, Urlaub H, Bottcher B, Szalay AA, Cramer P, Fischer U (2019): "Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes." Cell, 179, 1537-1550.e19. doi: 10.1016/j.cell.2019.11.024.
- Abstract
- Poxviruses encode a multisubunit DNA-dependent RNA polymerase (vRNAP) that carries out viral gene expression in the host cytoplasm. We report cryo-EM structures of core and complete vRNAP enzymes from Vaccinia virus at 2.8 Å resolution. The vRNAP core enzyme resembles eukaryotic RNA polymerase II (Pol II) but also reveals many virus-specific features, including the transcription factor Rap94. The complete enzyme additionally contains the transcription factor VETF, the mRNA processing factors VTF/CE and NPH-I, the viral core protein E11, and host tRNAGln. This complex can carry out the entire early transcription cycle. The structures show that Rap94 partially resembles the Pol II initiation factor TFIIB, that the vRNAP subunit Rpo30 resembles the Pol II elongation factor TFIIS, and that NPH-I resembles chromatin remodeling enzymes. Together with the accompanying paper (Hillen et al., 2019), these results provide the basis for unraveling the mechanisms of poxvirus transcription and RNA processing.