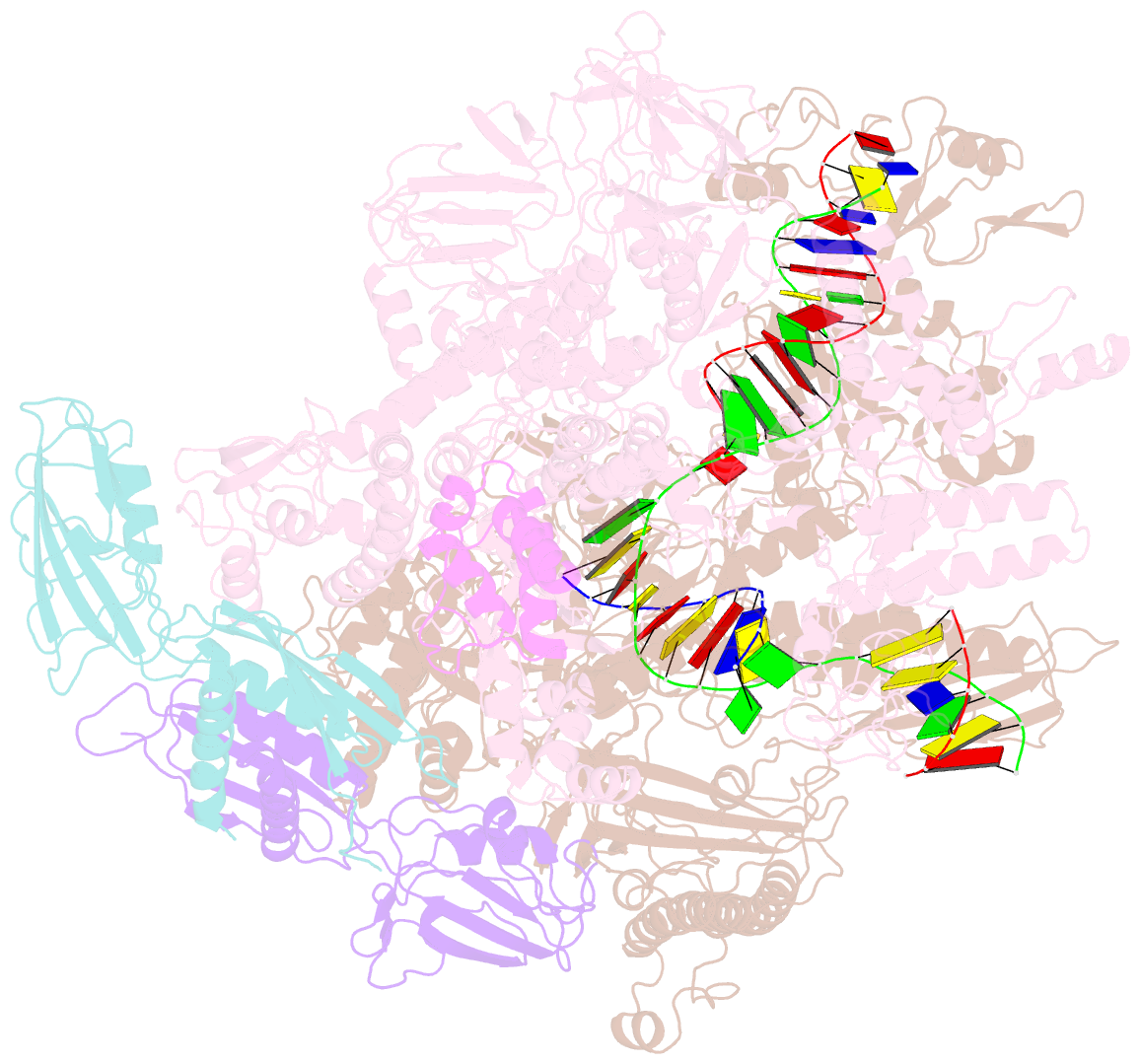
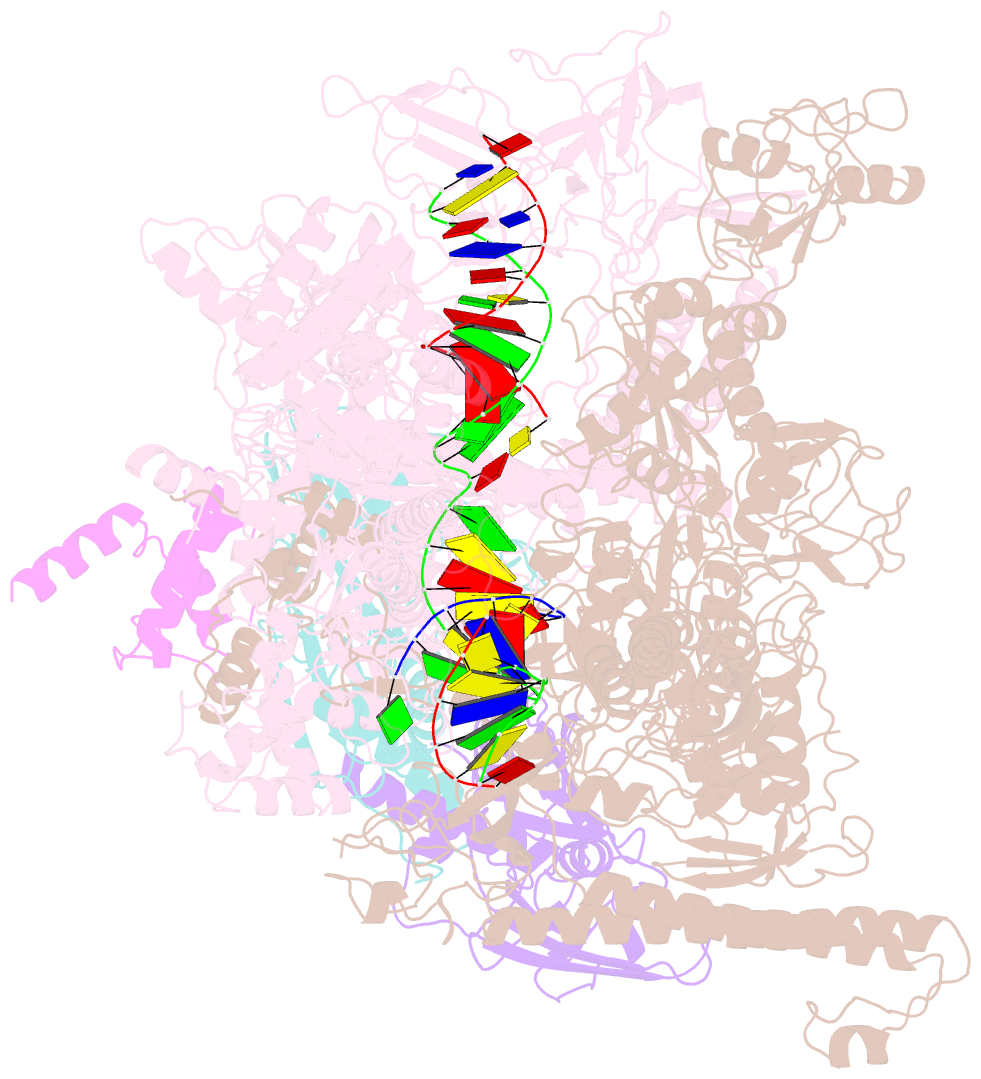
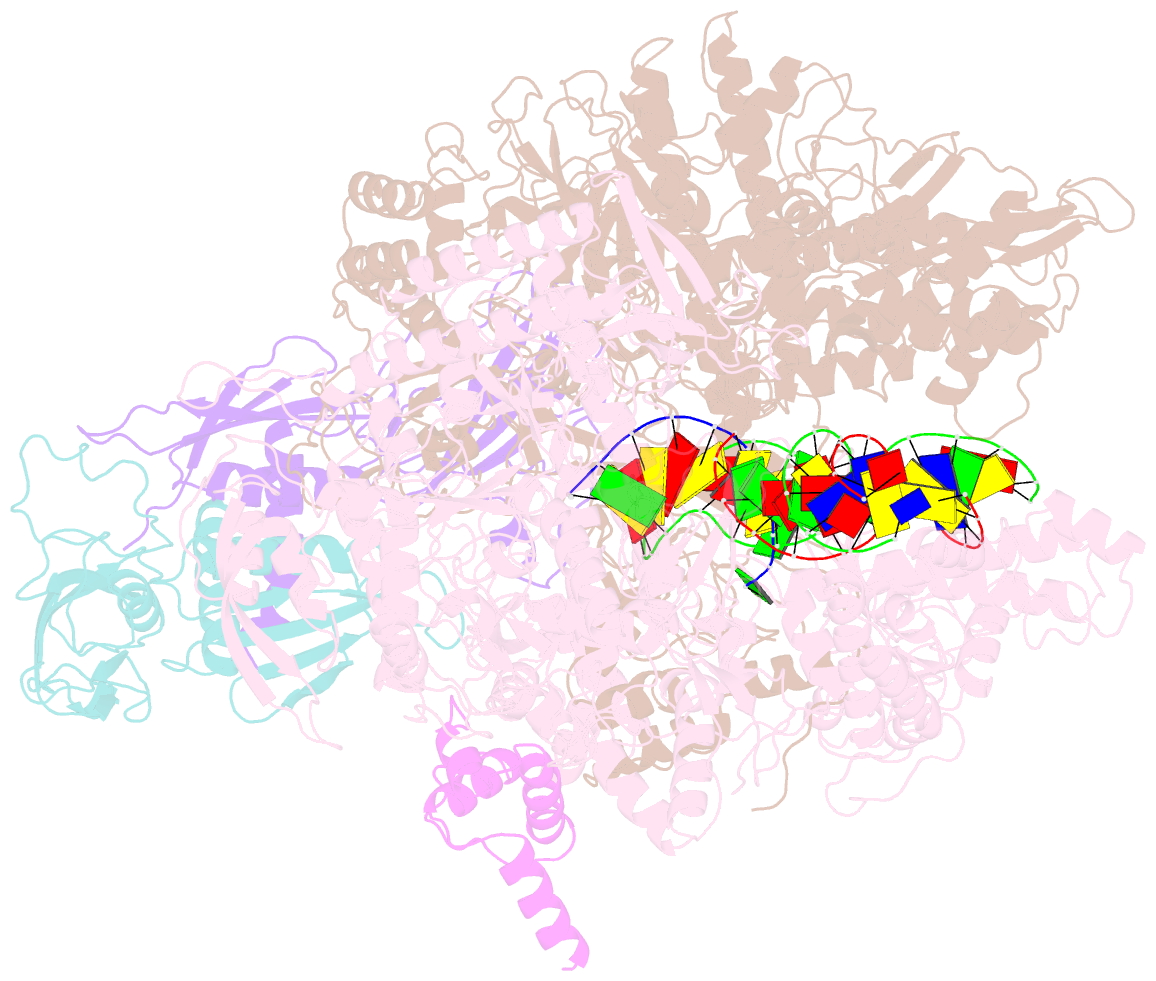
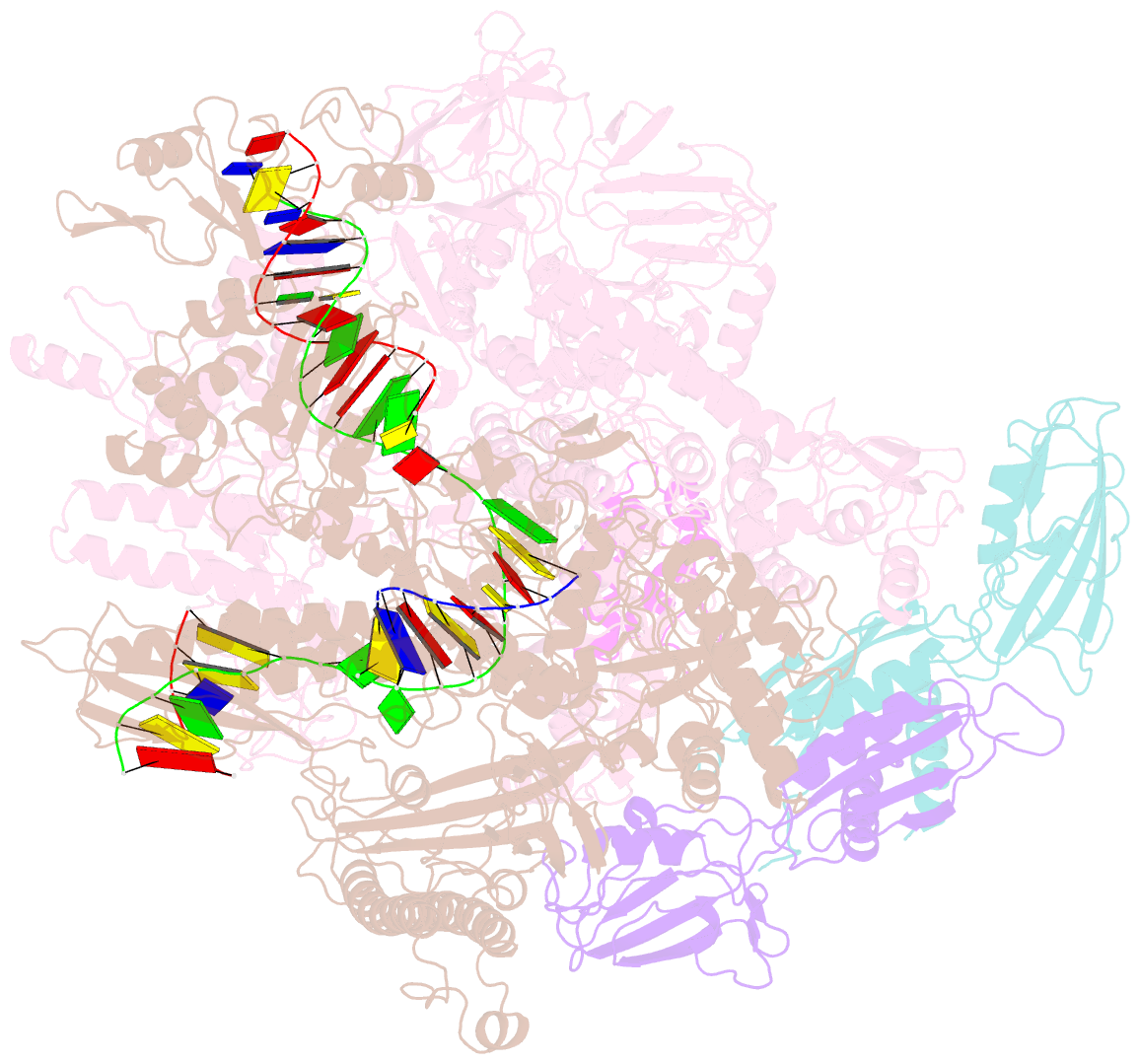
Summary information and primary citation
- PDB-id
- 6rh3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.6 Å)
- Summary
- cryo-EM structure of e. coli RNA polymerase elongation complex bound to ctp substrate
- Reference
- Abdelkareem M, Saint-Andre C, Takacs M, Papai G, Crucifix C, Guo X, Ortiz J, Weixlbaumer A (2019): "Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation." Mol.Cell, 75, 298-309.e4. doi: 10.1016/j.molcel.2019.04.029.
- Abstract
- Regulatory sequences or erroneous incorporations during DNA transcription cause RNA polymerase backtracking and inactivation in all kingdoms of life. Reactivation requires RNA transcript cleavage. Essential transcription factors (GreA and GreB, or TFIIS) accelerate this reaction. We report four cryo-EM reconstructions of Escherichia coli RNA polymerase representing the entire reaction pathway: (1) a backtracked complex; a backtracked complex with GreB (2) before and (3) after RNA cleavage; and (4) a reactivated, substrate-bound complex with GreB before RNA extension. Compared with eukaryotes, the backtracked RNA adopts a different conformation. RNA polymerase conformational changes cause distinct GreB states: a fully engaged GreB before cleavage; a disengaged GreB after cleavage; and a dislodged, loosely bound GreB removed from the active site to allow RNA extension. These reconstructions provide insight into the catalytic mechanism and dynamics of RNA cleavage and extension and suggest how GreB targets backtracked complexes without interfering with canonical transcription.