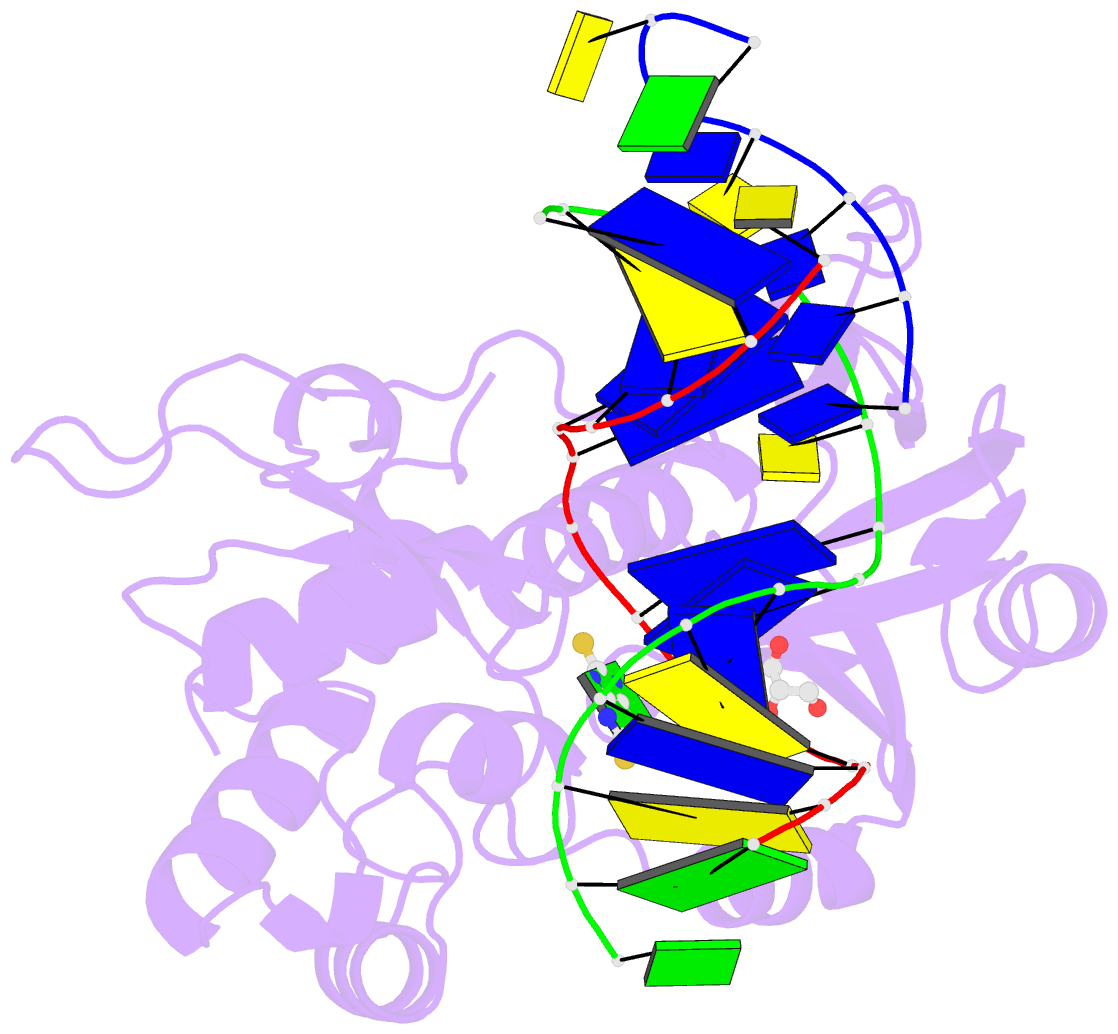
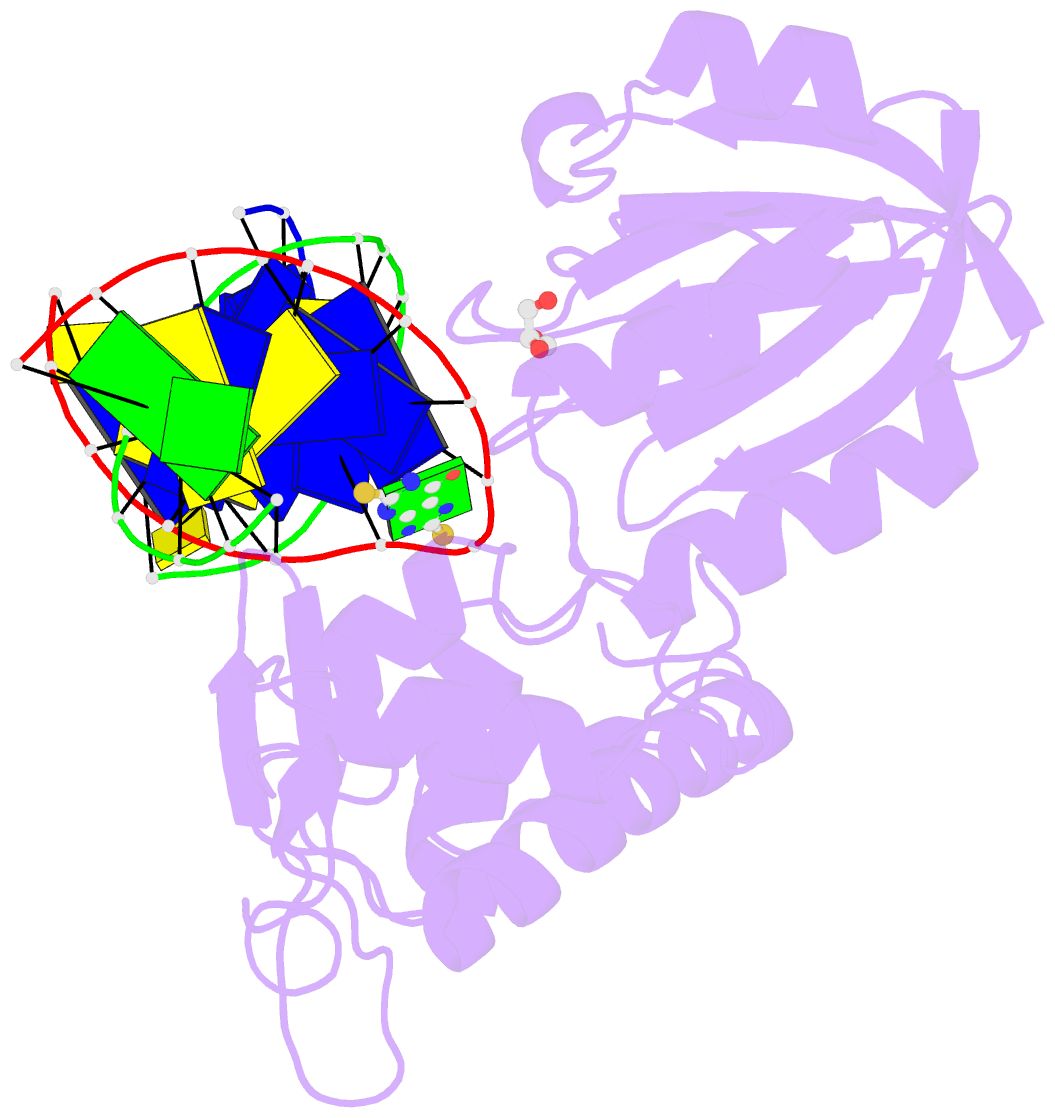
Summary information and primary citation
- PDB-id
- 6rp0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.25 Å)
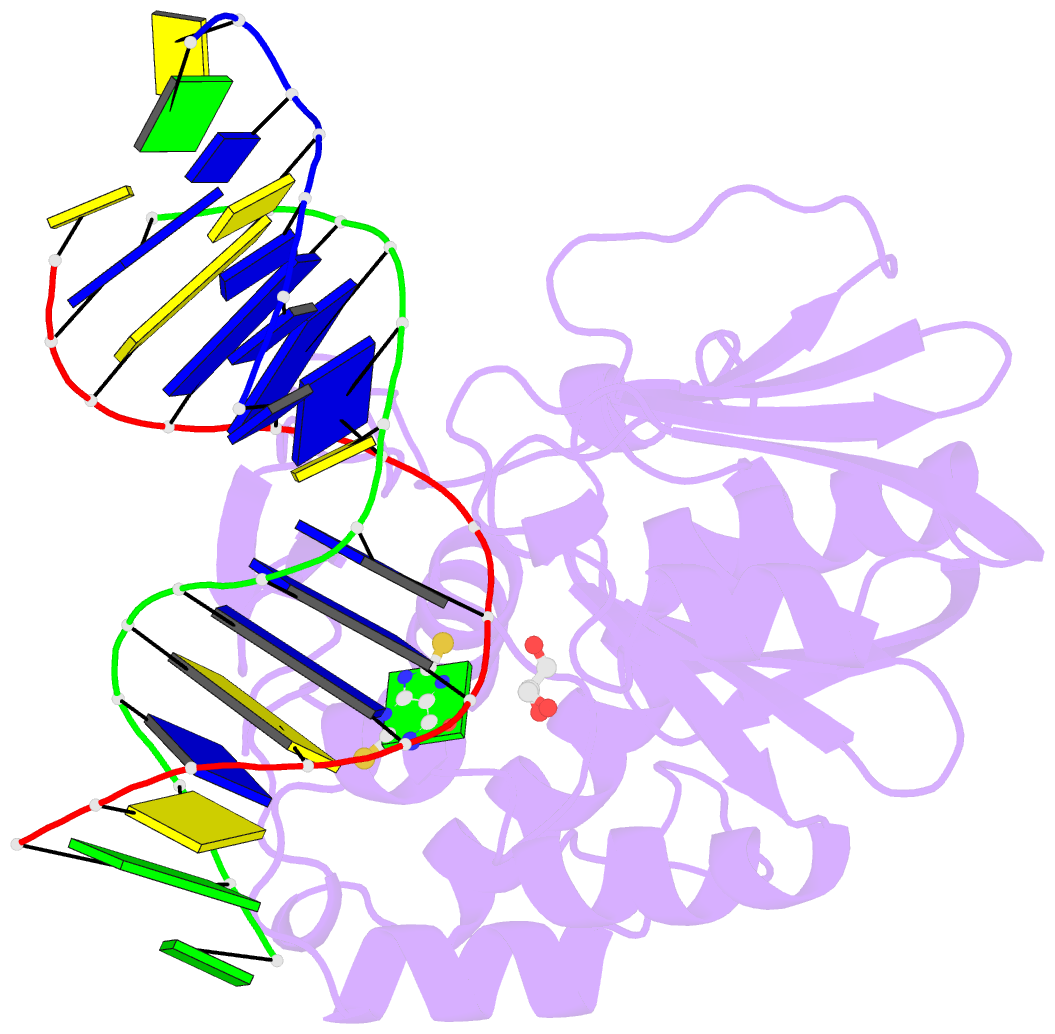
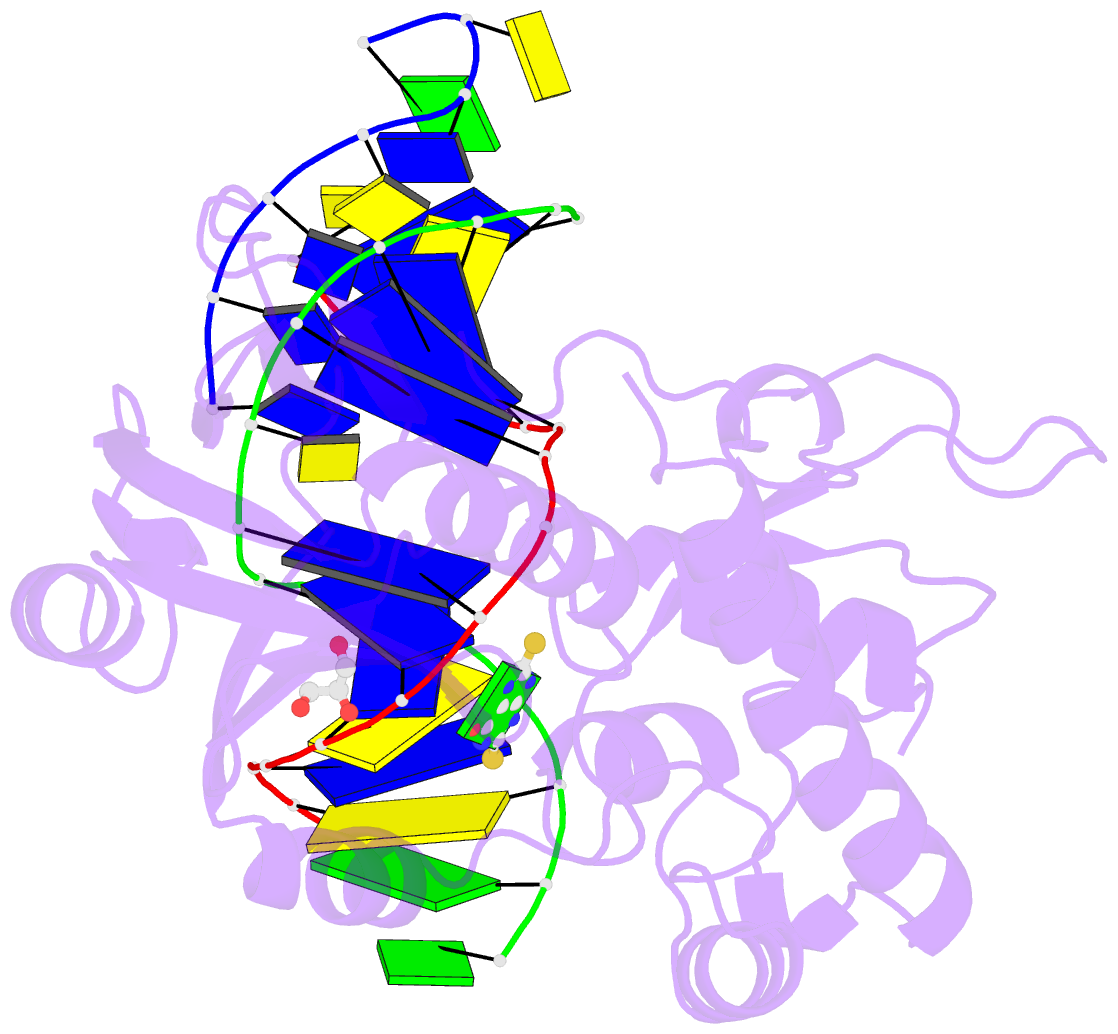
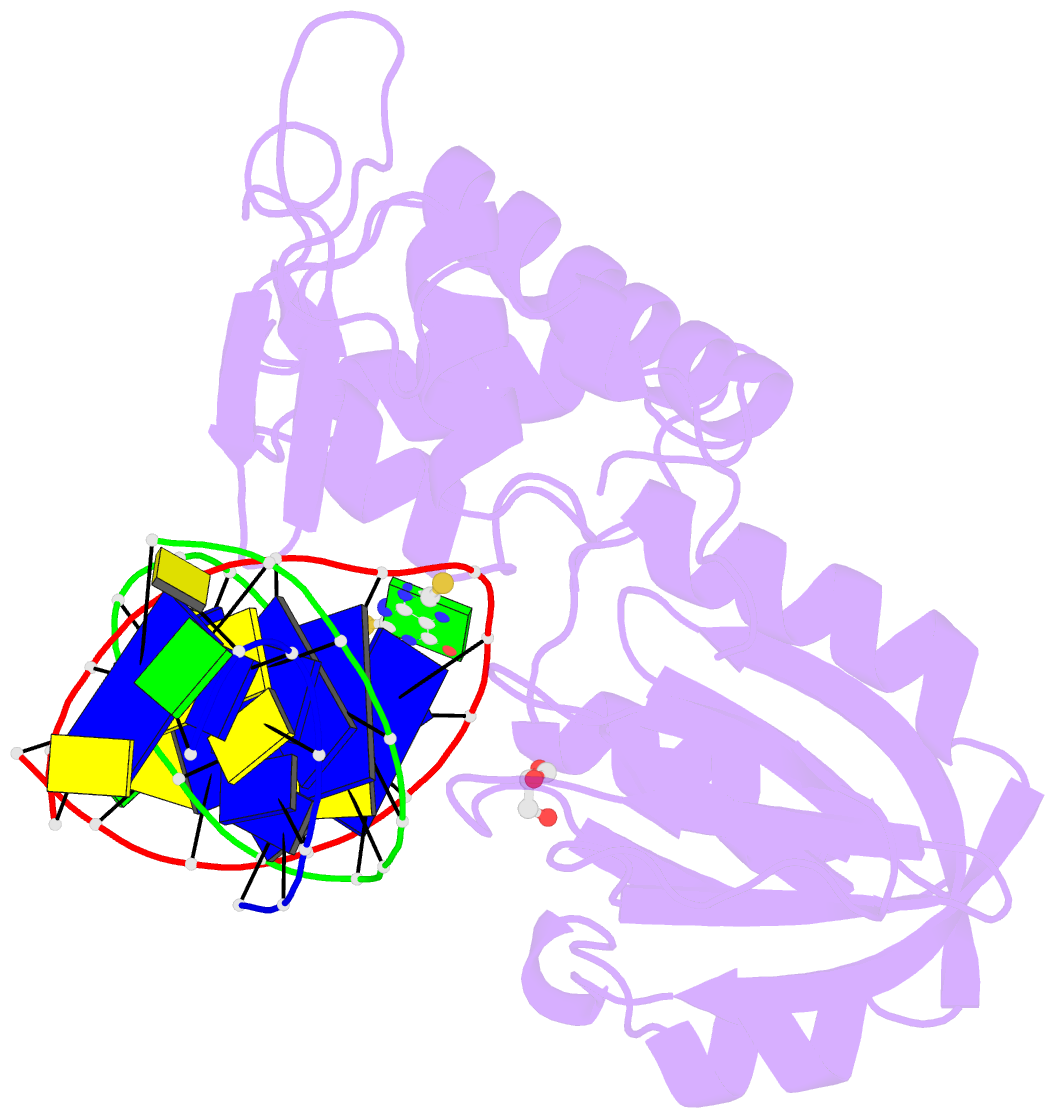
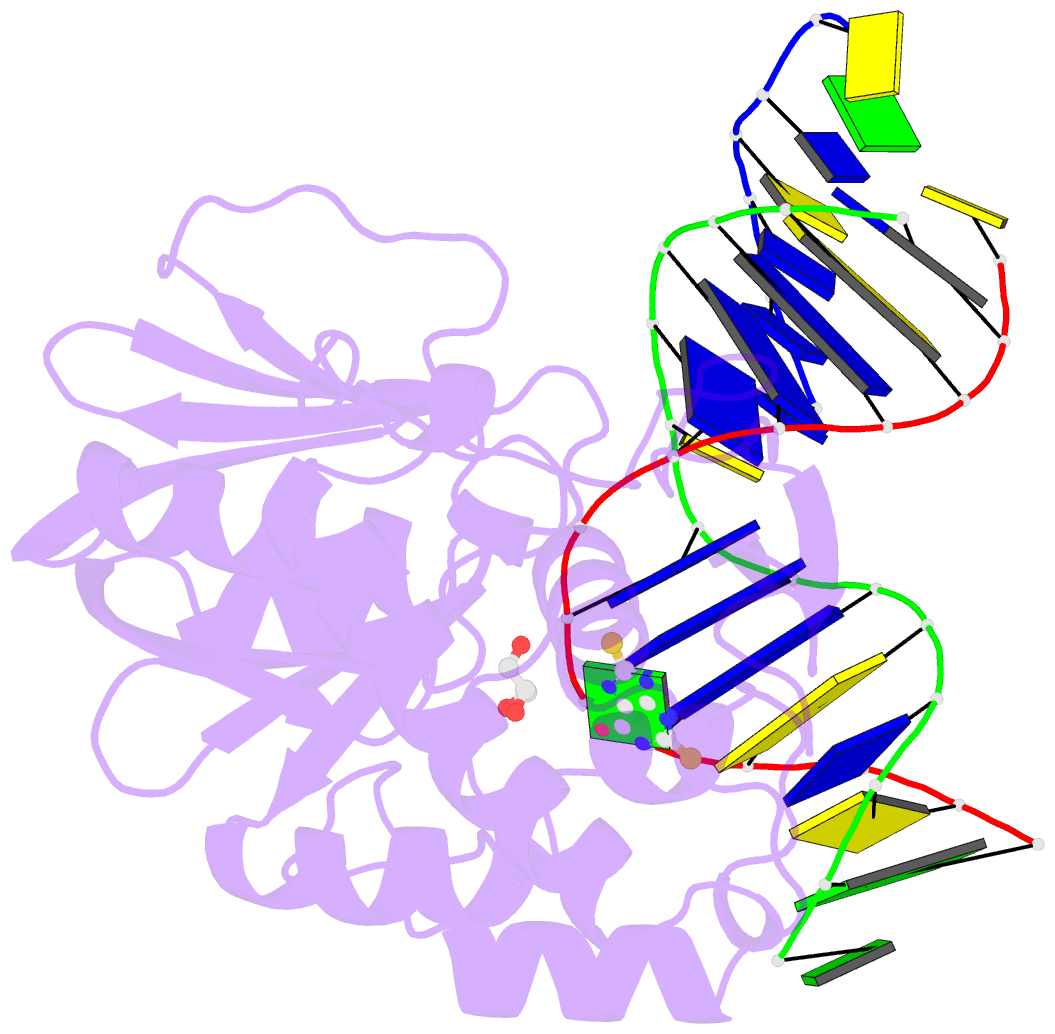
- Summary
- The crystal structure of a complex between the llfpg protein, a thf-DNA and an inhibitor
- Reference
- Rieux C, Goffinont S, Coste F, Tber Z, Cros J, Roy V, Guerin M, Gaudon V, Bourg S, Biela A, Aucagne V, Agrofoglio L, Garnier N, Castaing B (2020): "Thiopurine Derivative-Induced Fpg/Nei DNA Glycosylase Inhibition: Structural, Dynamic and Functional Insights." Int J Mol Sci, 21. doi: 10.3390/ijms21062058.
- Abstract
- DNA glycosylases are emerging as relevant pharmacological targets in inflammation, cancer and neurodegenerative diseases. Consequently, the search for inhibitors of these enzymes has become a very active research field. As a continuation of previous work that showed that 2-thioxanthine (2TX) is an irreversible inhibitor of zinc finger (ZnF)-containing Fpg/Nei DNA glycosylases, we designed and synthesized a mini-library of 2TX-derivatives (TXn) and evaluated their ability to inhibit Fpg/Nei enzymes. Among forty compounds, four TXn were better inhibitors than 2TX for Fpg. Unexpectedly, but very interestingly, two dithiolated derivatives more selectively and efficiently inhibit the zincless finger (ZnLF)-containing enzymes (human and mimivirus Neil1 DNA glycosylases hNeil1 and MvNei1, respectively). By combining chemistry, biochemistry, mass spectrometry, blind and flexible docking and X-ray structure analysis, we localized new TXn binding sites on Fpg/Nei enzymes. This endeavor allowed us to decipher at the atomic level the mode of action for the best TXn inhibitors on the ZnF-containing enzymes. We discovered an original inhibition mechanism for the ZnLF-containing Fpg/Nei DNA glycosylases by disulfide cyclic trimeric forms of dithiopurines. This work paves the way for the design and synthesis of a new structural class of inhibitors for selective pharmacological targeting of hNeil1 in cancer and neurodegenerative diseases.