Summary information and primary citation
- PDB-id
- 6s6h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.4 Å)
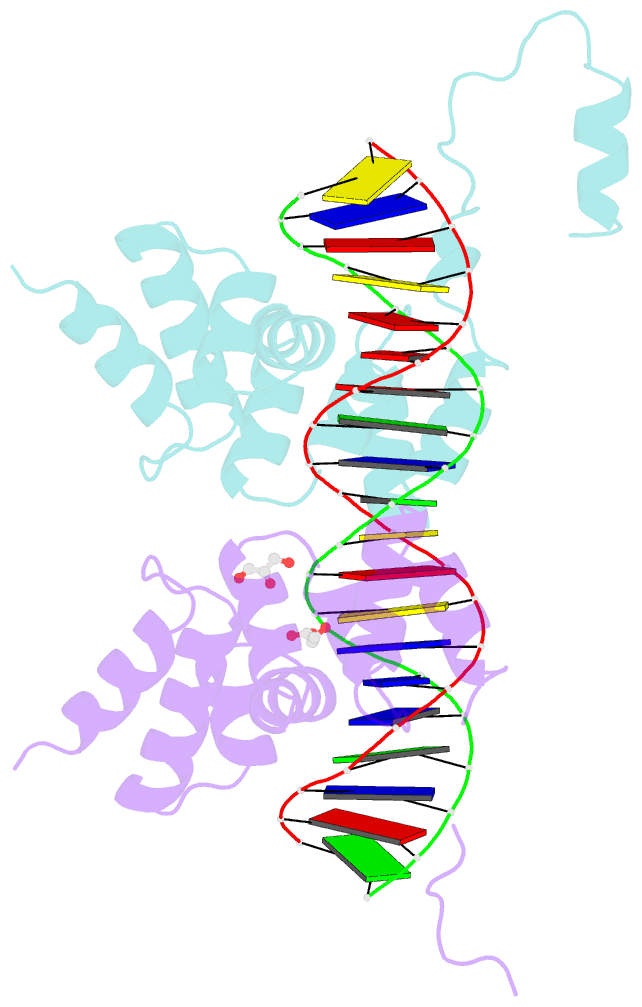
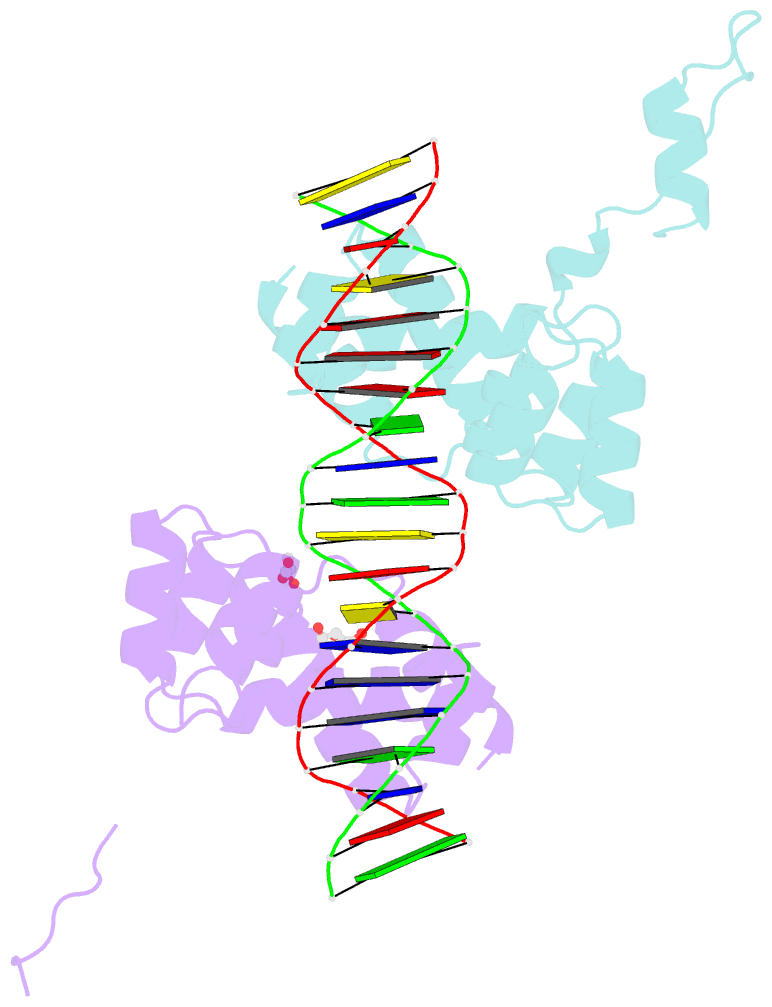
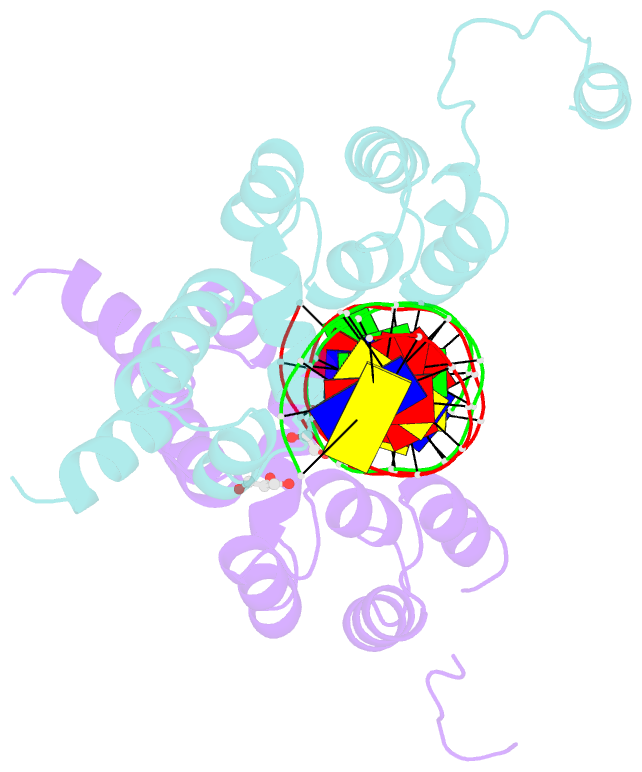
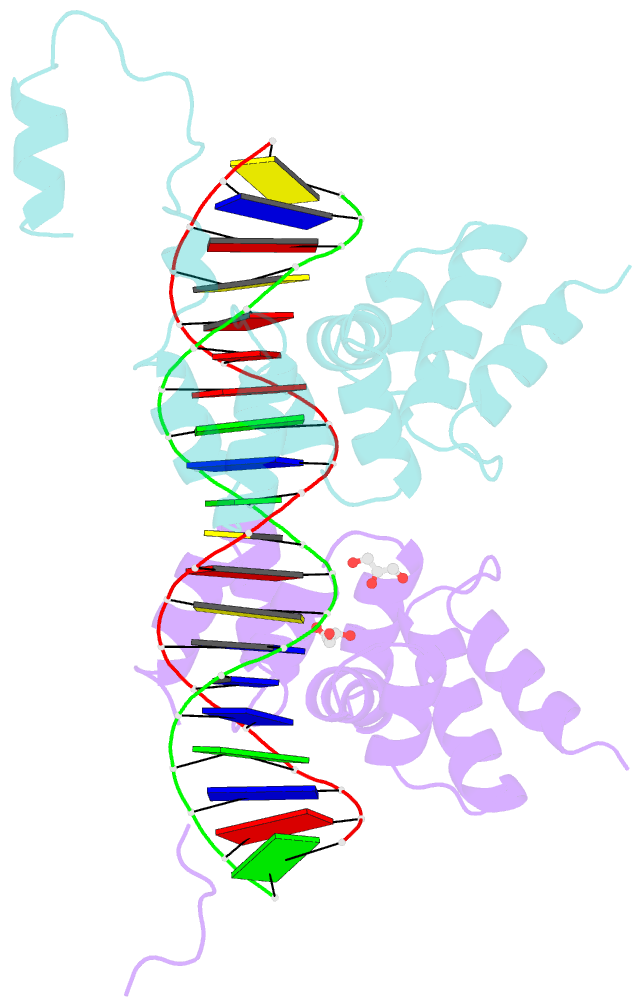
- Summary
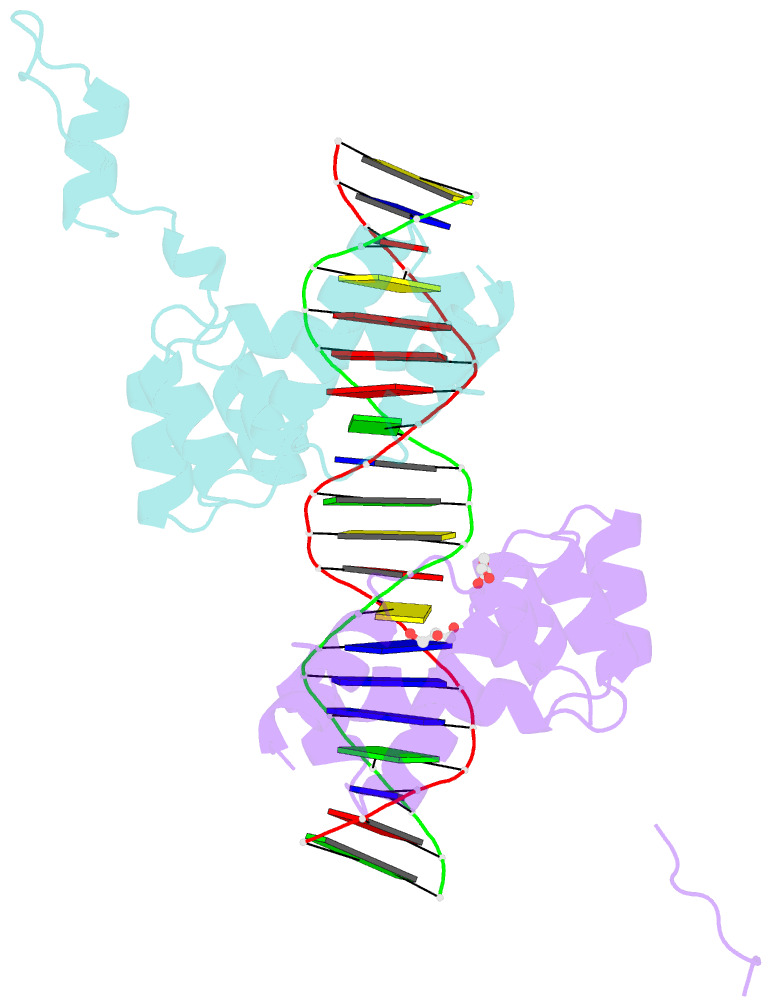
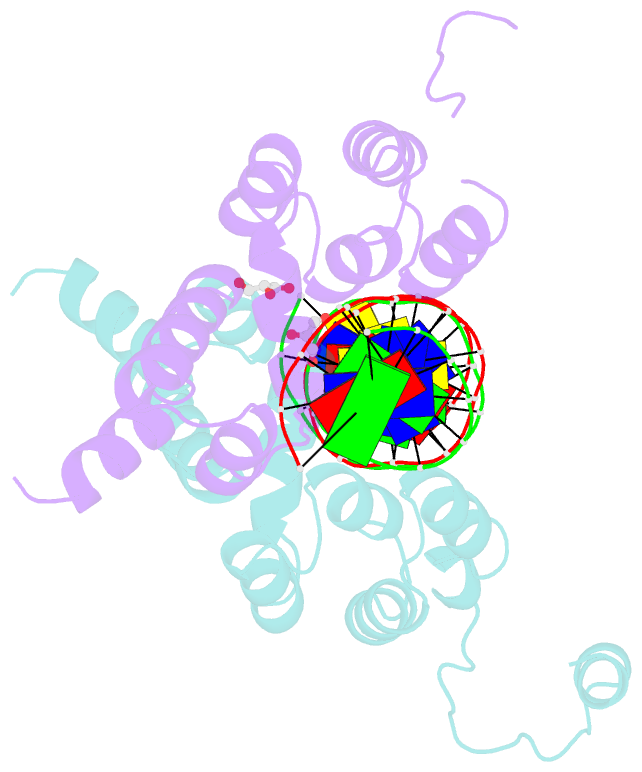
- Crystal structure of the DNA binding domain of the chromosome-partitioning protein parb complexed to the centromeric pars site
- Reference
- Jalal ASB, Tran NT, Stevenson CE, Chan EW, Lo R, Tan X, Noy A, Lawson DM, Le TBK (2020): "Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family." Cell Rep, 32, 107928. doi: 10.1016/j.celrep.2020.107928.
- Abstract
- Specific interactions between proteins and DNA are essential to many biological processes. Yet, it remains unclear how the diversification in DNA-binding specificity was brought about, and the mutational paths that led to changes in specificity are unknown. Using a pair of evolutionarily related DNA-binding proteins, each with a different DNA preference (ParB [Partitioning Protein B] and Noc [Nucleoid Occlusion Factor], which both play roles in bacterial chromosome maintenance), we show that specificity is encoded by a set of four residues at the protein-DNA interface. Combining X-ray crystallography and deep mutational scanning of the interface, we suggest that permissive mutations must be introduced before specificity-switching mutations to reprogram specificity and that mutational paths to new specificity do not necessarily involve dual-specificity intermediates. Overall, our results provide insight into the possible evolutionary history of ParB and Noc and, in a broader context, might be useful for understanding the evolution of other classes of DNA-binding proteins.