Summary information and primary citation
- PDB-id
- 6sce; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA
- Method
- X-ray (1.83 Å)
- Summary
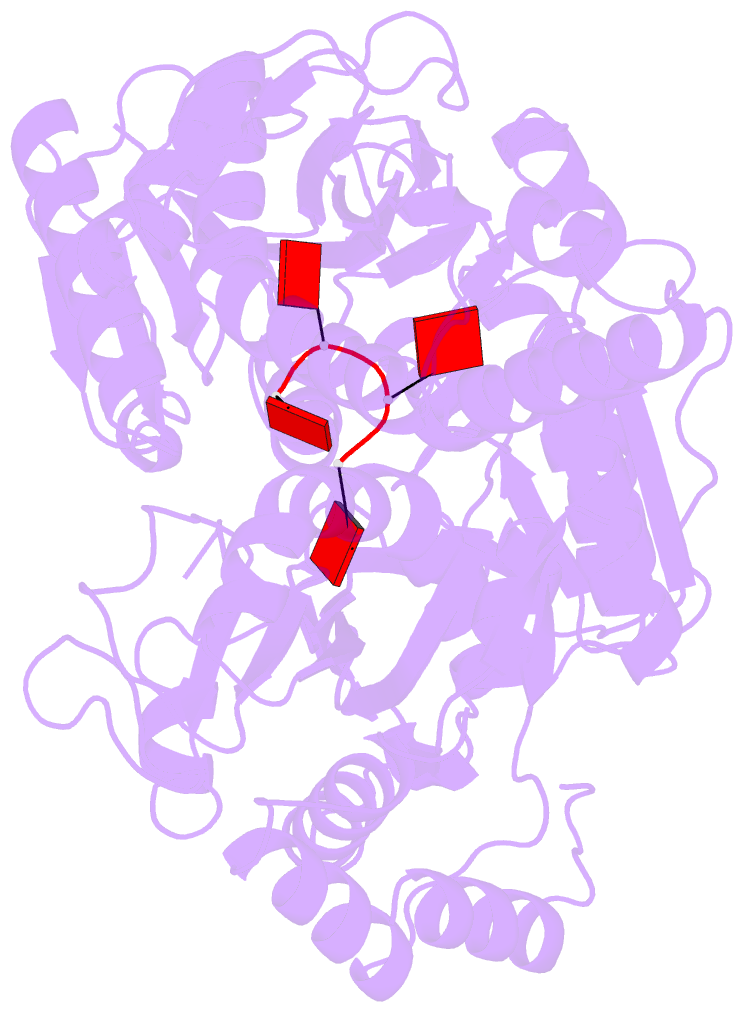
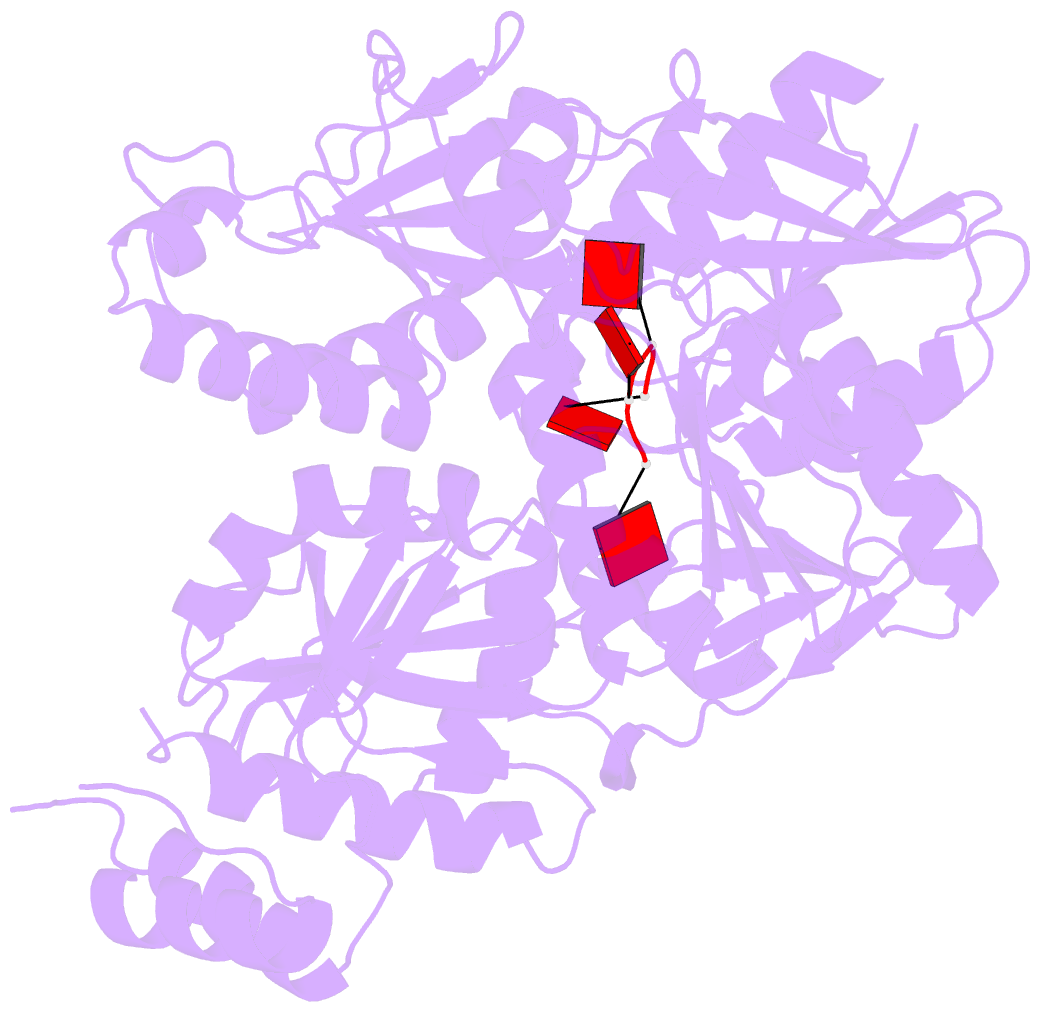
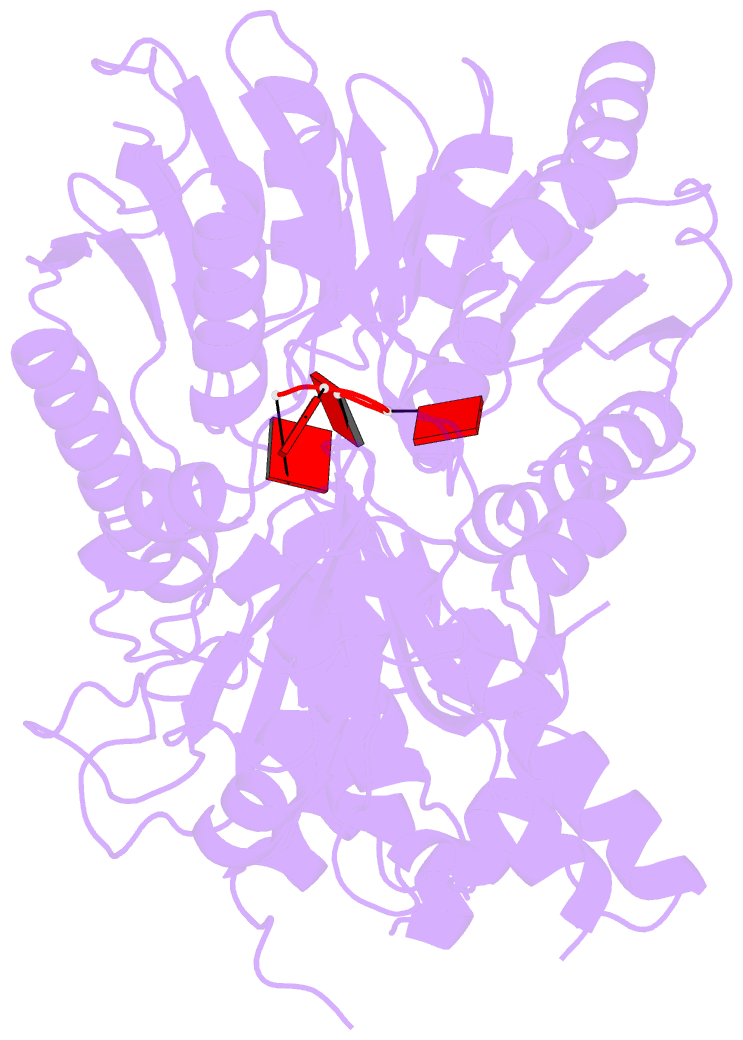
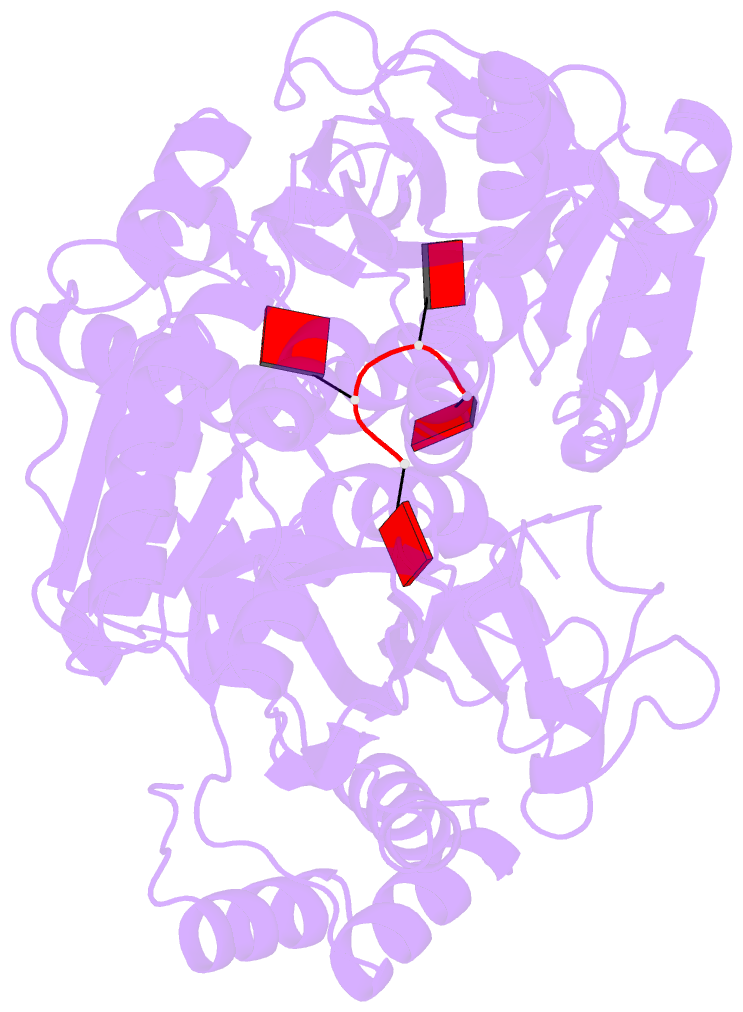
- Structure of a type iii crispr defence DNA nuclease activated by cyclic oligoadenylate
- Reference
- McMahon SA, Zhu W, Graham S, Rambo R, White MF, Gloster TM (2020): "Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate." Nat Commun, 11, 500. doi: 10.1038/s41467-019-14222-x.
- Abstract
- The CRISPR system provides adaptive immunity against mobile genetic elements in prokaryotes. On binding invading RNA species, Type III CRISPR systems generate cyclic oligoadenylate (cOA) signalling molecules, potentiating a powerful immune response by activating downstream effector proteins, leading to viral clearance, cell dormancy or death. Here we describe the structure and mechanism of a cOA-activated CRISPR defence DNA endonuclease, CRISPR ancillary nuclease 1 (Can1). Can1 has a unique monomeric structure with two CRISPR associated Rossman fold (CARF) domains and two DNA nuclease-like domains. The crystal structure of the enzyme has been captured in the activated state, with a cyclic tetra-adenylate (cA4) molecule bound at the core of the protein. cA4 binding reorganises the structure to license a metal-dependent DNA nuclease activity specific for nicking of supercoiled DNA. DNA nicking by Can1 is predicted to slow down viral replication kinetics by leading to the collapse of DNA replication forks.