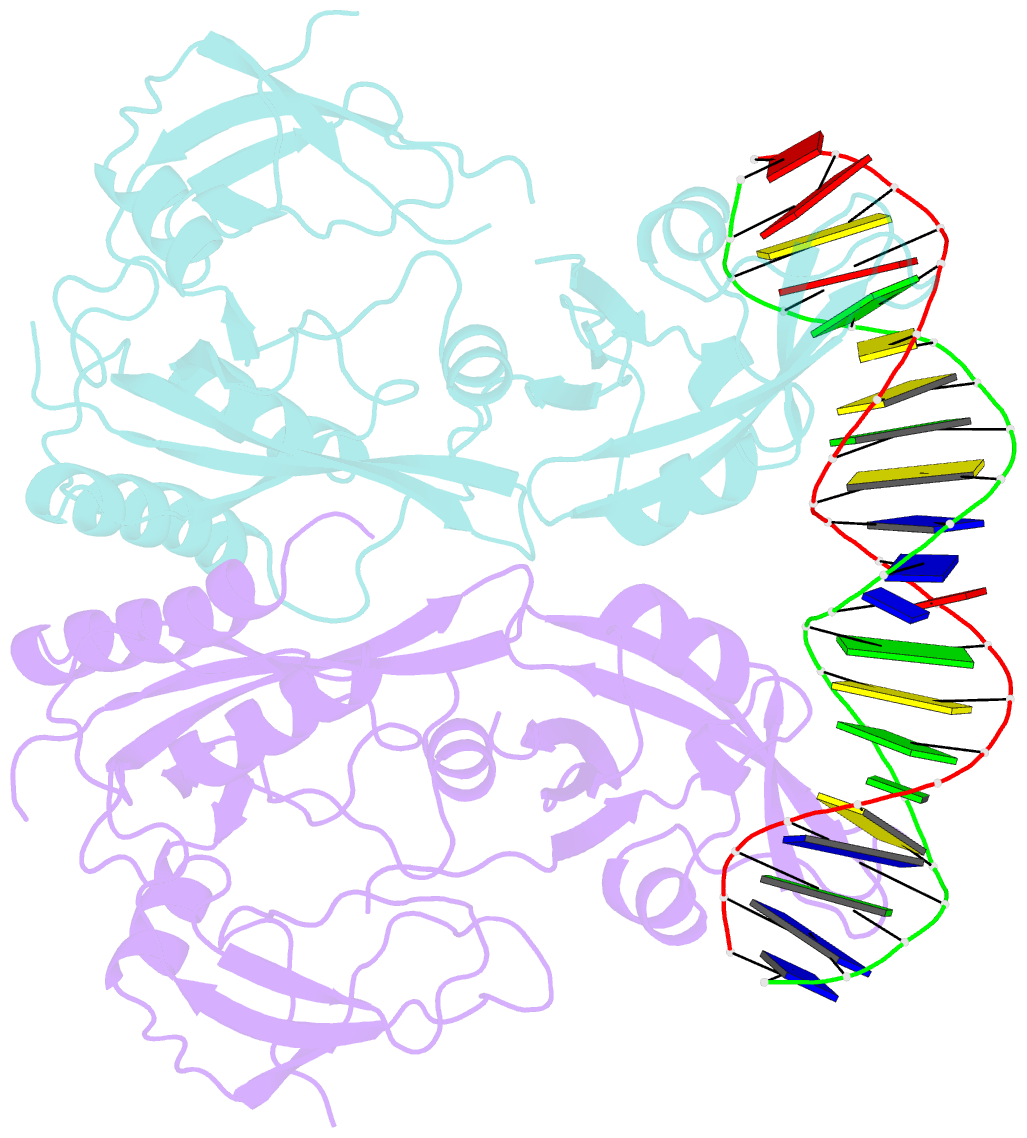
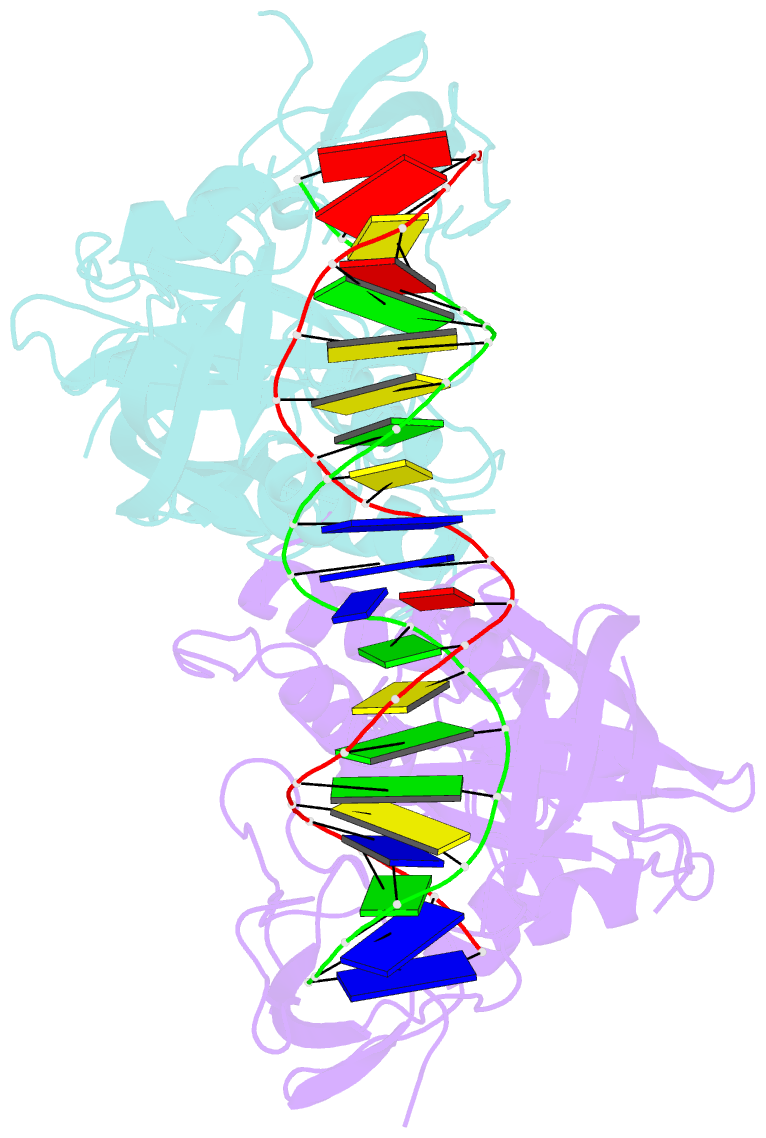
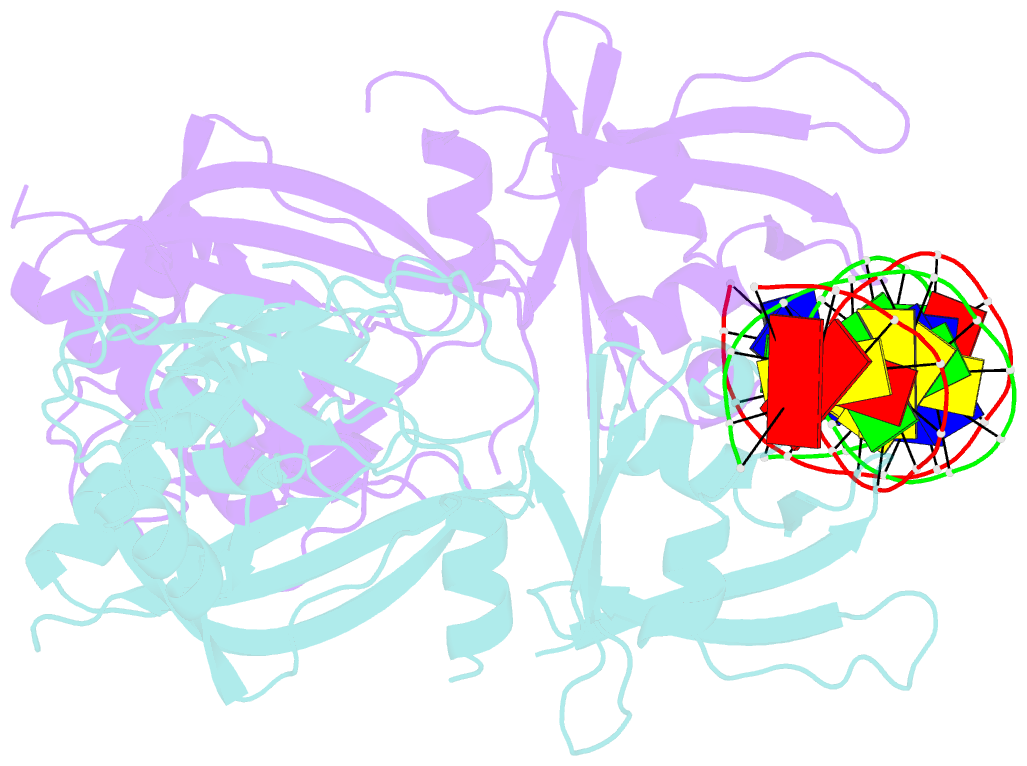
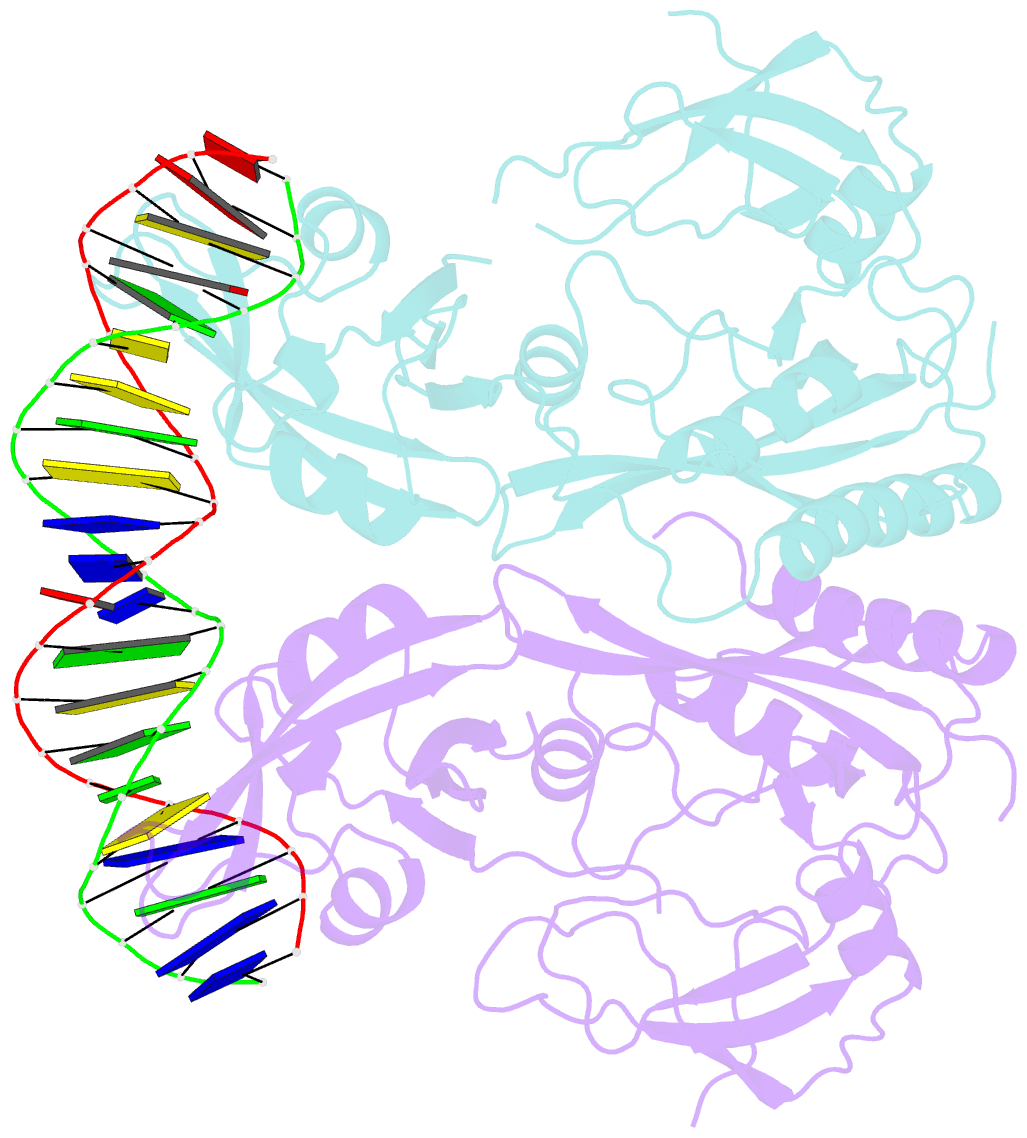
Summary information and primary citation
- PDB-id
- 6sdg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.96 Å)
- Summary
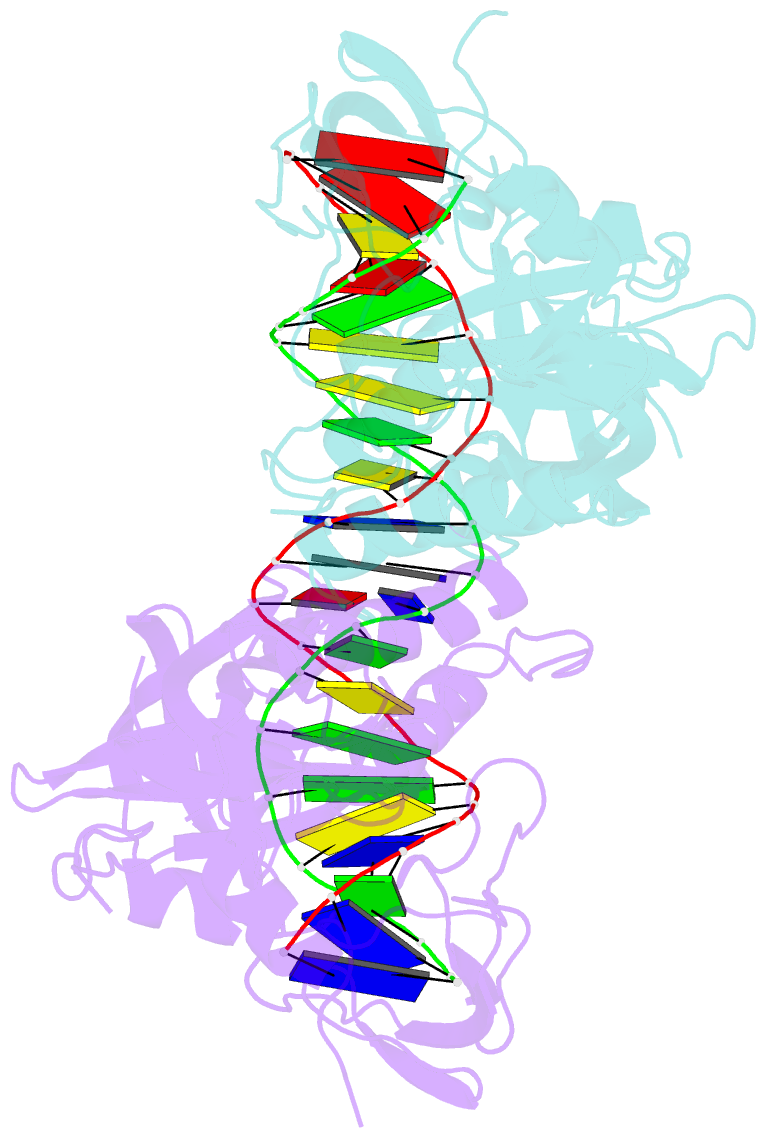
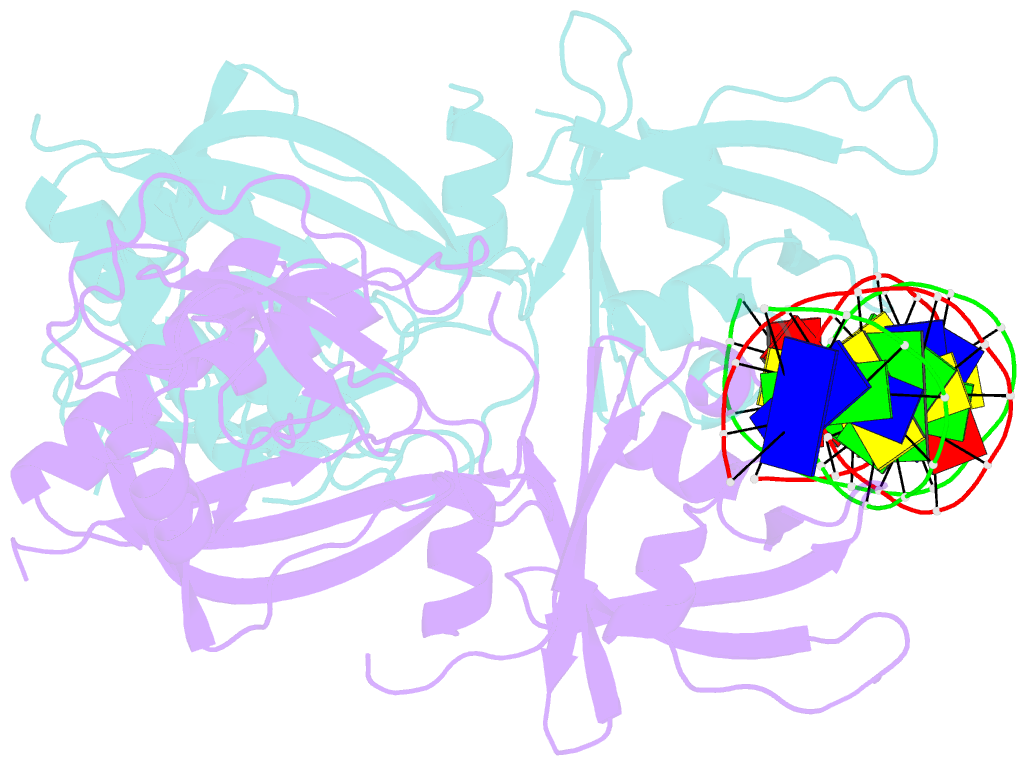
- Crystal structure of the DNA binding domain of m. polymorpha auxin response factor 2 (mparf2) in complex with high affinity DNA
- Reference
- Kato H, Mutte SK, Suzuki H, Crespo I, Das S, Radoeva T, Fontana M, Yoshitake Y, Hainiwa E, van den Berg W, Lindhoud S, Ishizaki K, Hohlbein J, Borst JW, Boer DR, Nishihama R, Kohchi T, Weijers D (2020): "Design principles of a minimal auxin response system." Nat.Plants, 6, 473-482. doi: 10.1038/s41477-020-0662-y.
- Abstract
- Auxin controls numerous growth processes in land plants through a gene expression system that modulates ARF transcription factor activity1-3. Gene duplications in families encoding auxin response components have generated tremendous complexity in most land plants, and neofunctionalization enabled various unique response outputs during development1,3,4. However, it is unclear what fundamental biochemical principles underlie this complex response system. By studying the minimal system in Marchantia polymorpha, we derive an intuitive and simple model where a single auxin-dependent A-ARF activates gene expression. It is antagonized by an auxin-independent B-ARF that represses common target genes. The expression patterns of both ARF proteins define developmental zones where auxin response is permitted, quantitatively tuned or prevented. This fundamental design probably represents the ancestral system and formed the basis for inflated, complex systems.