Summary information and primary citation
- PDB-id
- 6sx0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- X-ray (1.75 Å)
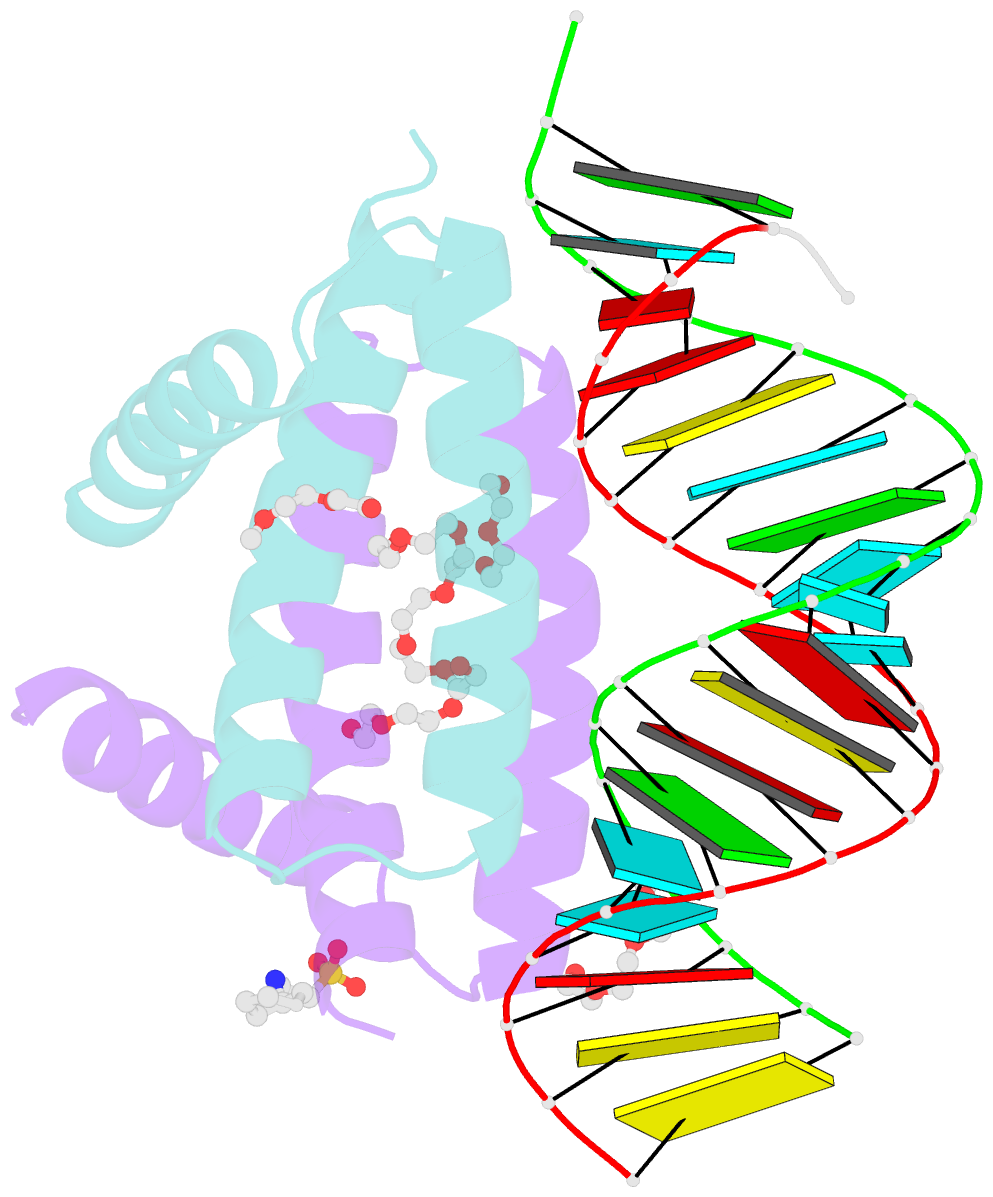
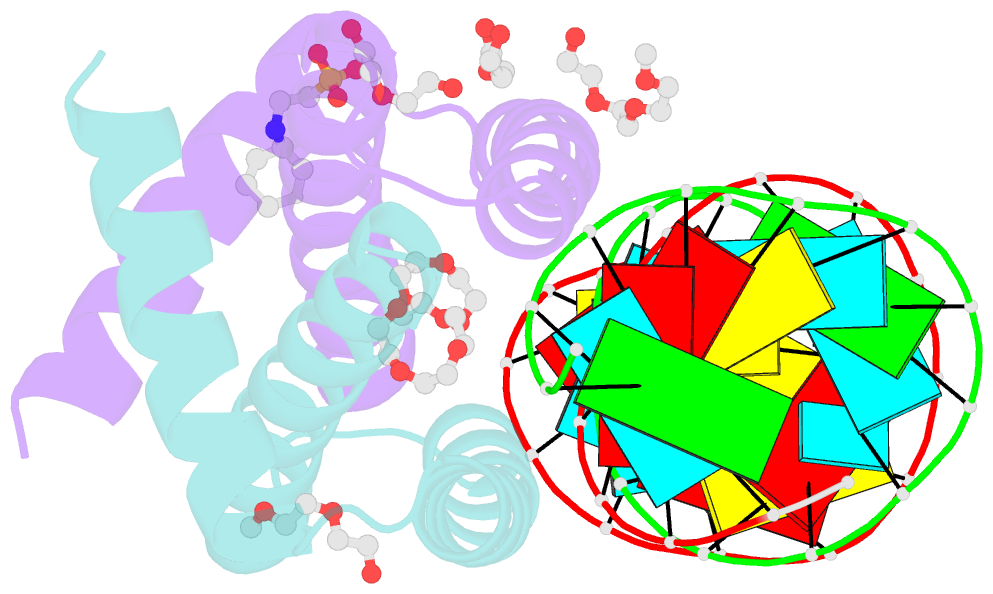
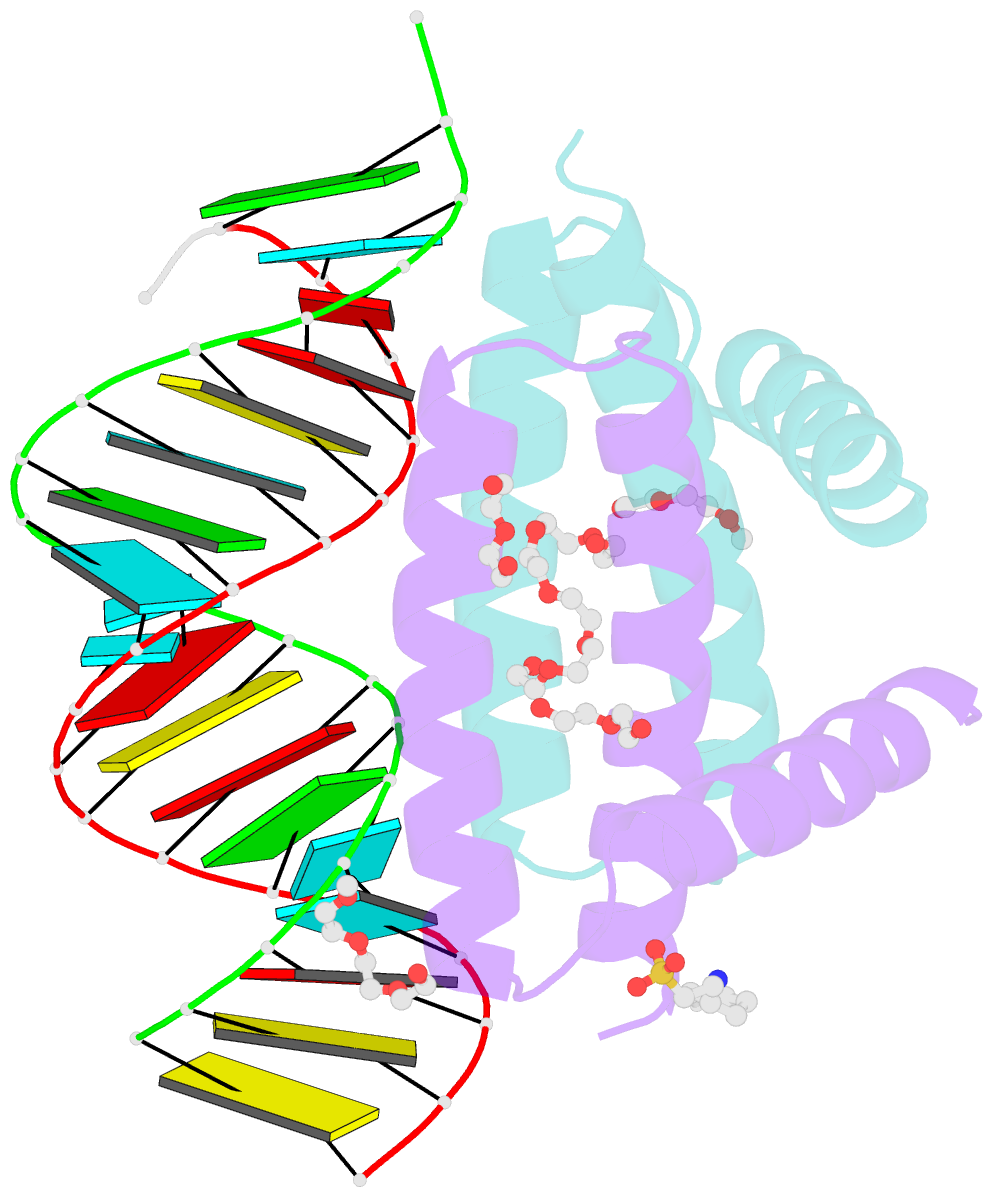
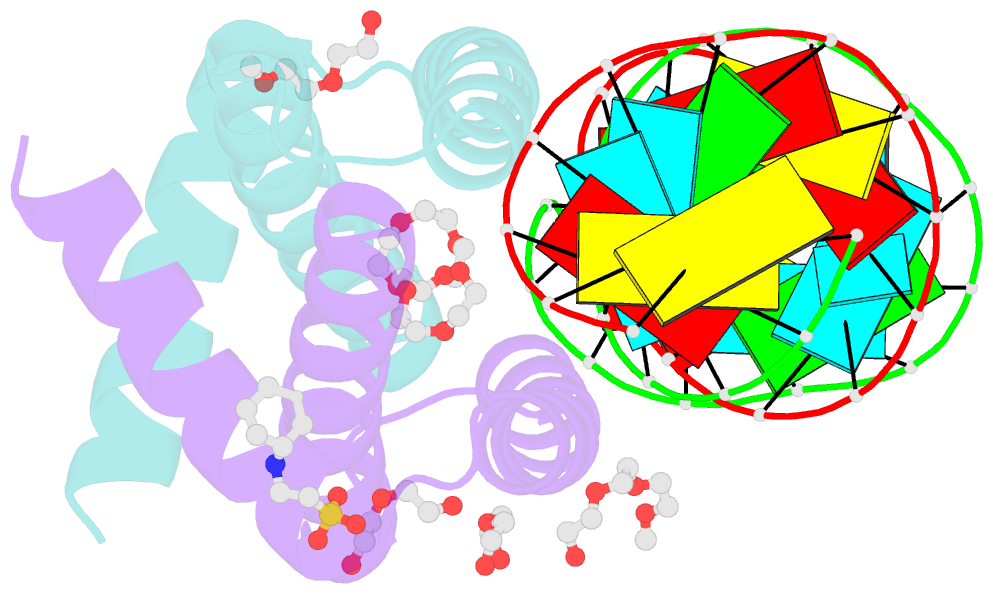
- Summary
- Specific dsrna recognition by wild type h7n1 ns1 RNA-binding domain
- Reference
- Wacquiez A, Coste F, Kut E, Gaudon V, Trapp S, Castaing B, Marc D (2020): "Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1." Viruses, 12. doi: 10.3390/v12090947.
- Abstract
- The non-structural protein NS1 of influenza A viruses is an RNA-binding protein of which its activities in the infected cell contribute to the success of the viral cycle, notably through interferon antagonism. We have previously shown that NS1 strongly binds RNA aptamers harbouring virus-specific sequence motifs (Marc et al., Nucleic Acids Res. 41, 434-449). Here, we started out investigating the putative role of one particular virus-specific motif through the phenotypic characterization of mutant viruses that were genetically engineered from the parental strain WSN. Unexpectedly, our data did not evidence biological importance of the putative binding of NS1 to this specific motif (UGAUUGAAG) in the 3'-untranslated region of its own mRNA. Next, we sought to identify specificity determinants in the NS1-RNA interaction through interaction assays in vitro with several RNA ligands and through solving by X-ray diffraction the 3D structure of several complexes associating NS1's RBD with RNAs of various affinities. Our data show that the RBD binds the GUAAC motif within double-stranded RNA helices with an apparent specificity that may rely on the sequence-encoded ability of the RNA to bend its axis. On the other hand, we showed that the RBD binds to the virus-specific AGCAAAAG motif when it is exposed in the apical loop of a high-affinity RNA aptamer, probably through a distinct mode of interaction that still requires structural characterization. Our data are consistent with more than one mode of interaction of NS1's RBD with RNAs, recognizing both structure and sequence determinants.