Summary information and primary citation
- PDB-id
- 6t34; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- virus
- Method
- cryo-EM (5.2 Å)
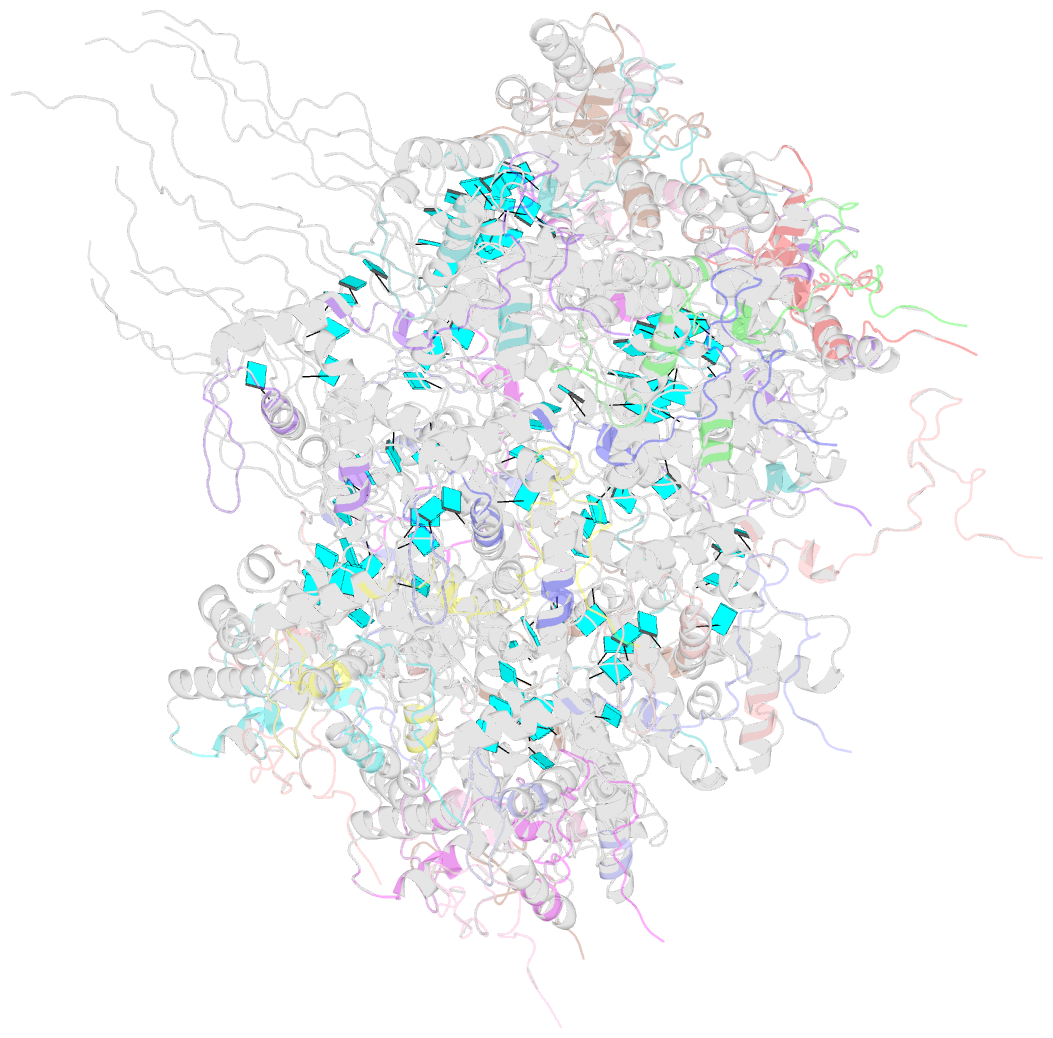
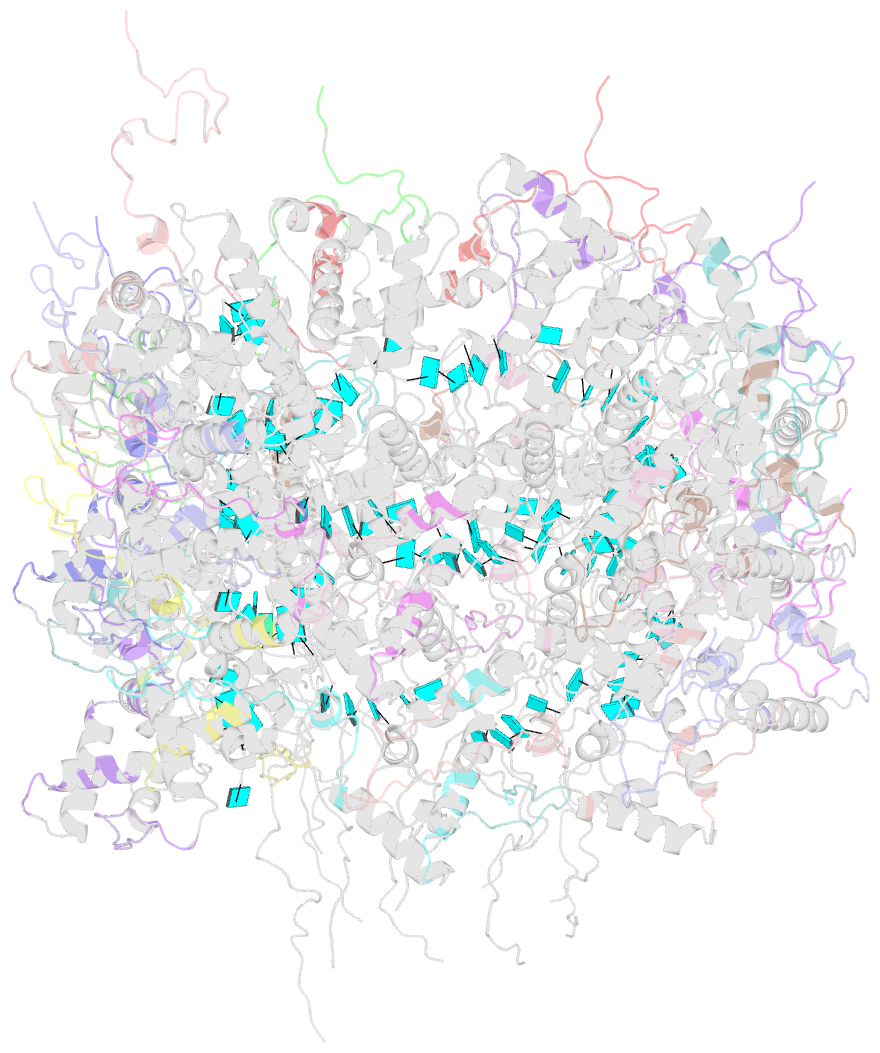
- Summary
- Atomic model for turnip mosaic virus (tumv)
- Reference
- Cuesta R, Yuste-Calvo C, Gil-Carton D, Sanchez F, Ponz F, Valle M (2019): "Structure of Turnip mosaic virus and its viral-like particles." Sci Rep, 9, 15396. doi: 10.1038/s41598-019-51823-4.
- Abstract
- Turnip mosaic virus (TuMV), a potyvirus, is a flexible filamentous plant virus that displays a helical arrangement of coat protein copies (CPs) bound to the ssRNA genome. TuMV is a bona fide representative of the Potyvirus genus, one of most abundant groups of plant viruses, which displays a very wide host range. We have studied by cryoEM the structure of TuMV virions and its viral-like particles (VLPs) to explore the role of the interactions between proteins and RNA in the assembly of the virions. The results show that the CP-RNA interaction is needed for the correct orientation of the CP N-terminal arm, a region that plays as a molecular staple between CP subunits in the fully assembled virion.