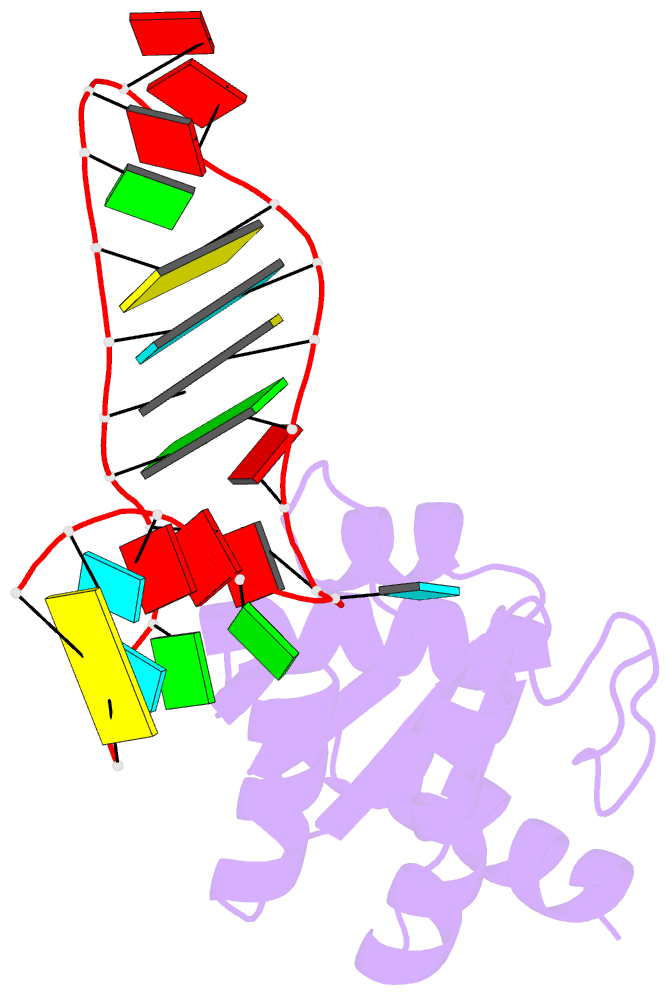
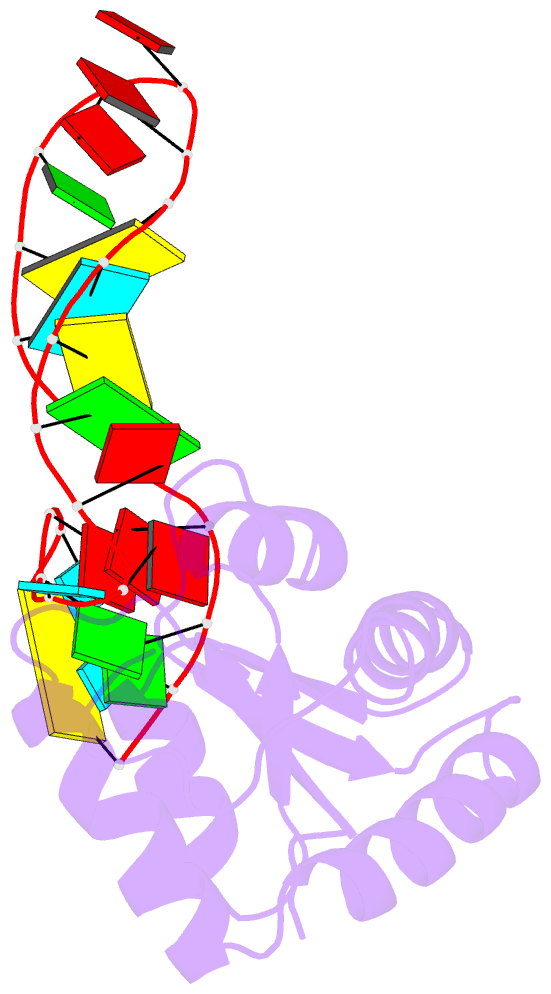
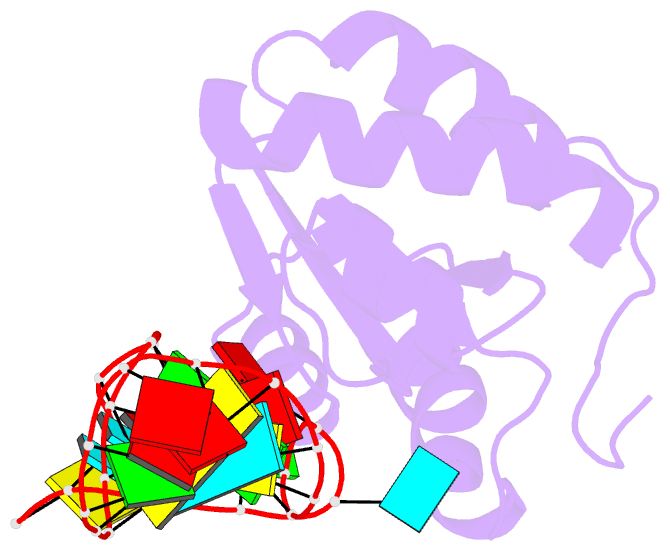
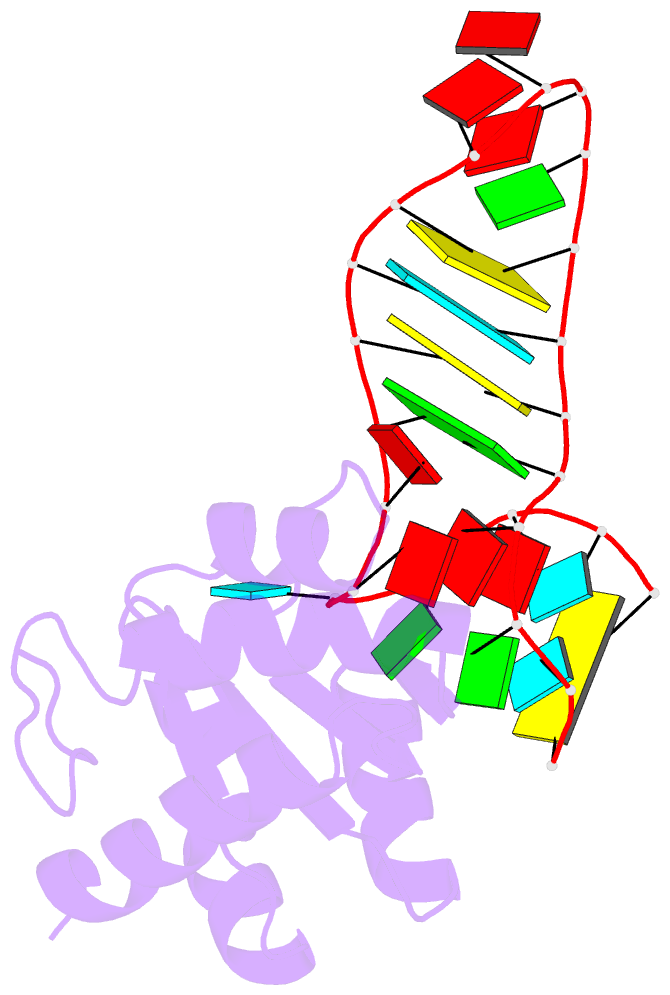
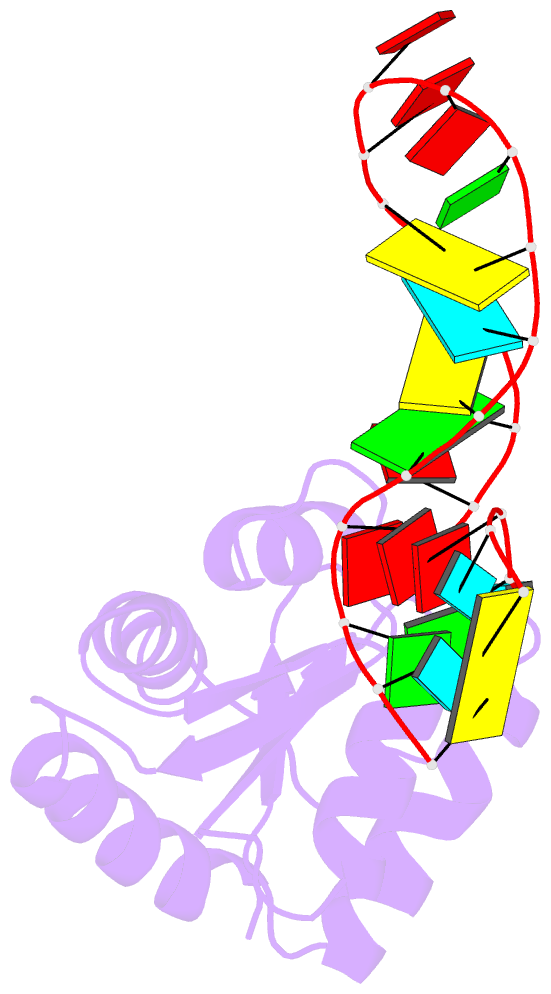
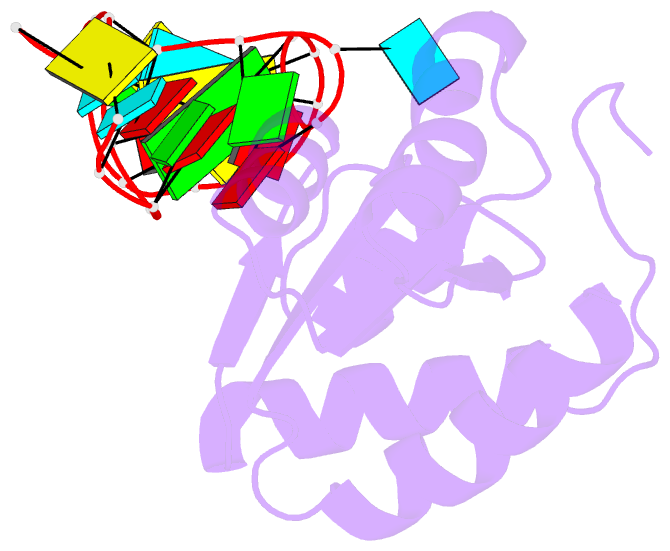
Summary information and primary citation
- PDB-id
- 6tph; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA
- Method
- NMR
- Summary
- Structure of a protein-RNA complex by ssnmr
- Reference
- Ahmed M, Marchanka A, Carlomagno T (2020): "Structure of a Protein-RNA Complex by Solid-State NMR Spectroscopy." Angew.Chem.Int.Ed.Engl., 59, 6866-6873. doi: 10.1002/anie.201915465.
- Abstract
- Solid-state NMR (ssNMR) is applicable to high molecular-weight (MW) protein assemblies in a non-amorphous precipitate. The technique yields atomic resolution structural information on both soluble and insoluble particles without limitations of MW or requirement of crystals. Herein, we propose and demonstrate an approach that yields the structure of protein-RNA complexes (RNP) solely from ssNMR data. Instead of using low-sensitivity magnetization transfer steps between heteronuclei of the protein and the RNA, we measure paramagnetic relaxation enhancement effects elicited on the RNA by a paramagnetic tag coupled to the protein. We demonstrate that this data, together with chemical-shift-perturbation data, yields an accurate structure of an RNP complex, starting from the bound structures of its components. The possibility of characterizing protein-RNA interactions by ssNMR may enable applications to large RNP complexes, whose structures are not accessible by other methods.