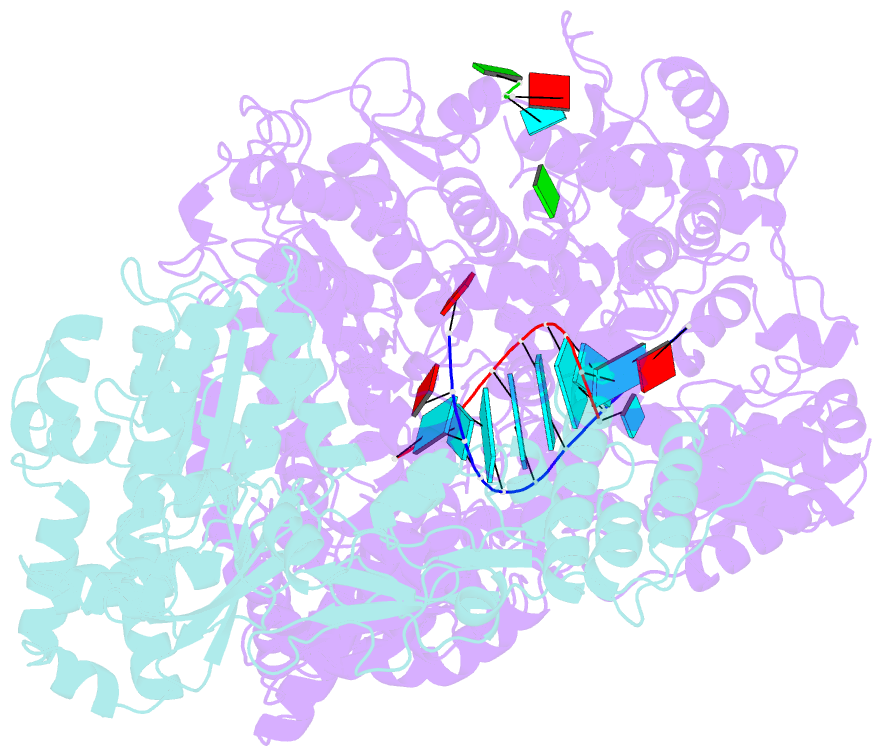
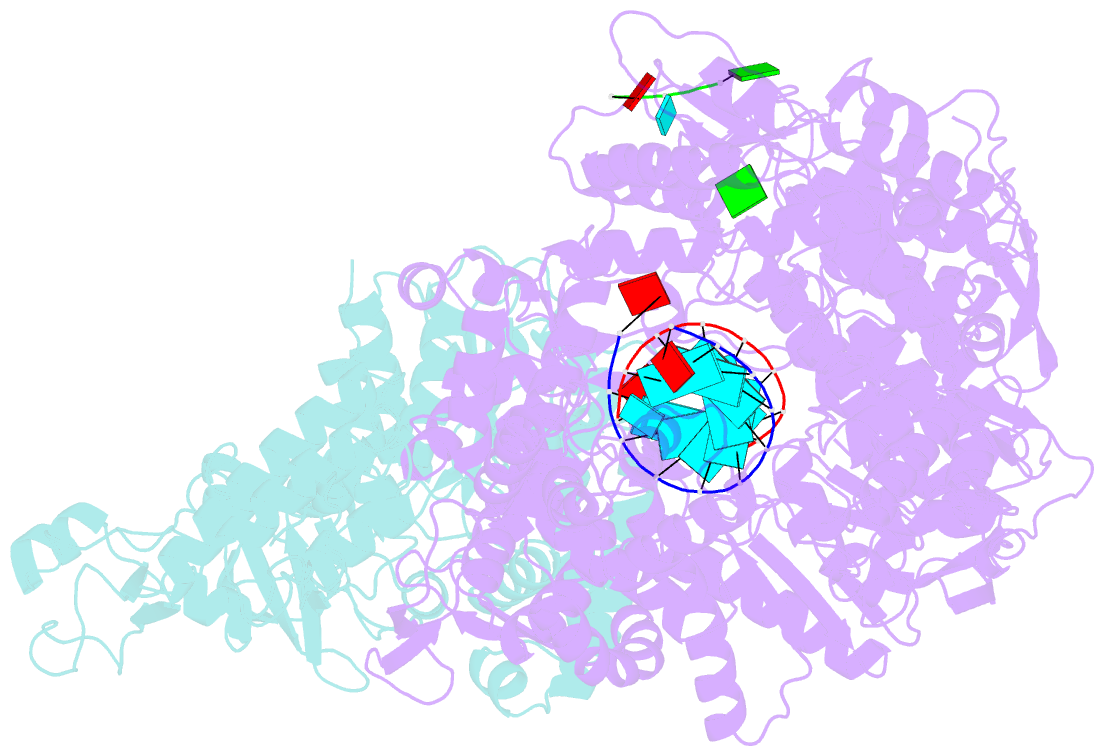
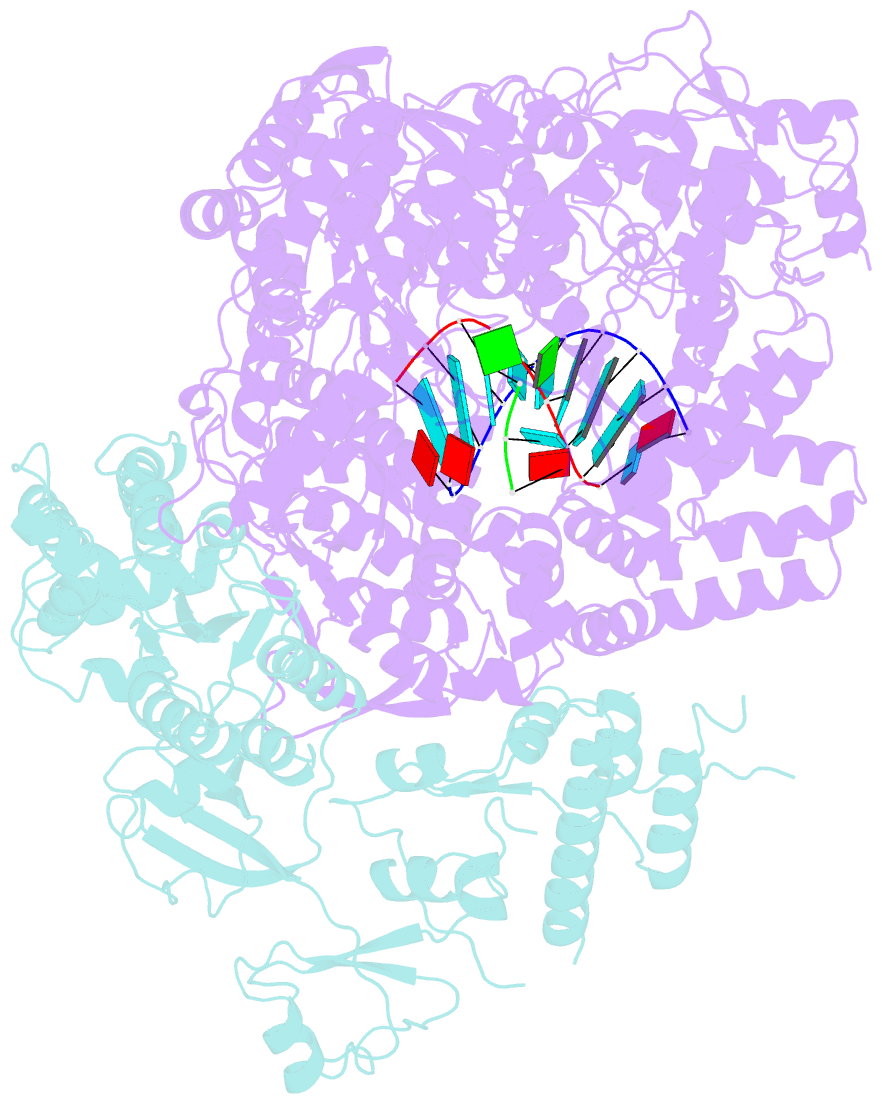
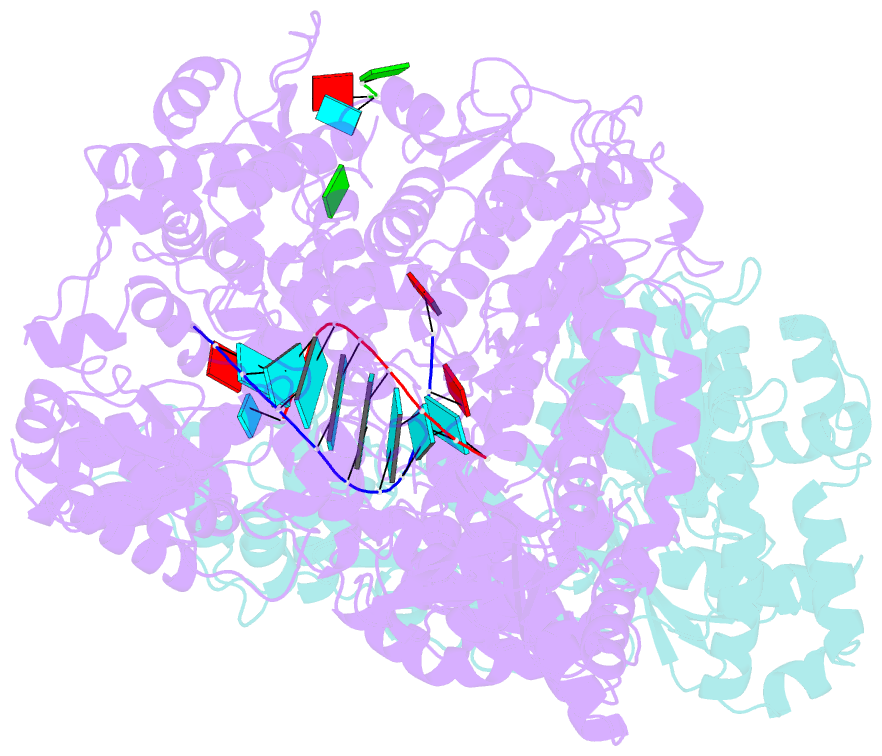
Summary information and primary citation
- PDB-id
- 6tz1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein, transferase-RNA
- Method
- cryo-EM (3.4 Å)
- Summary
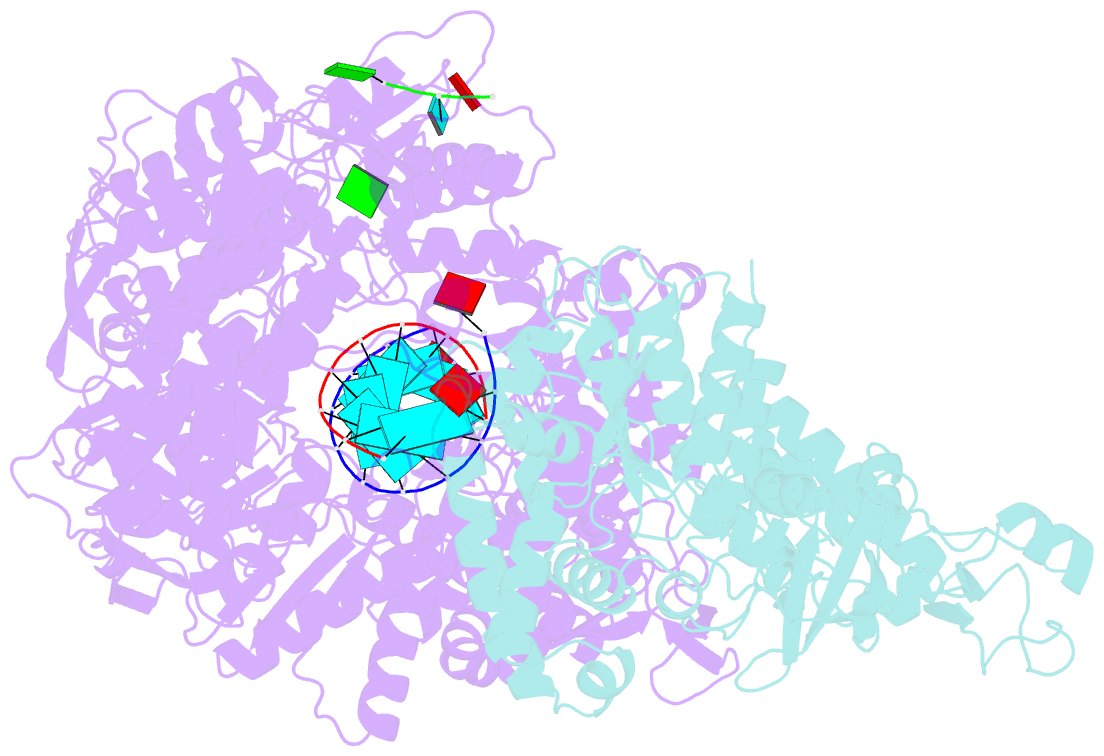
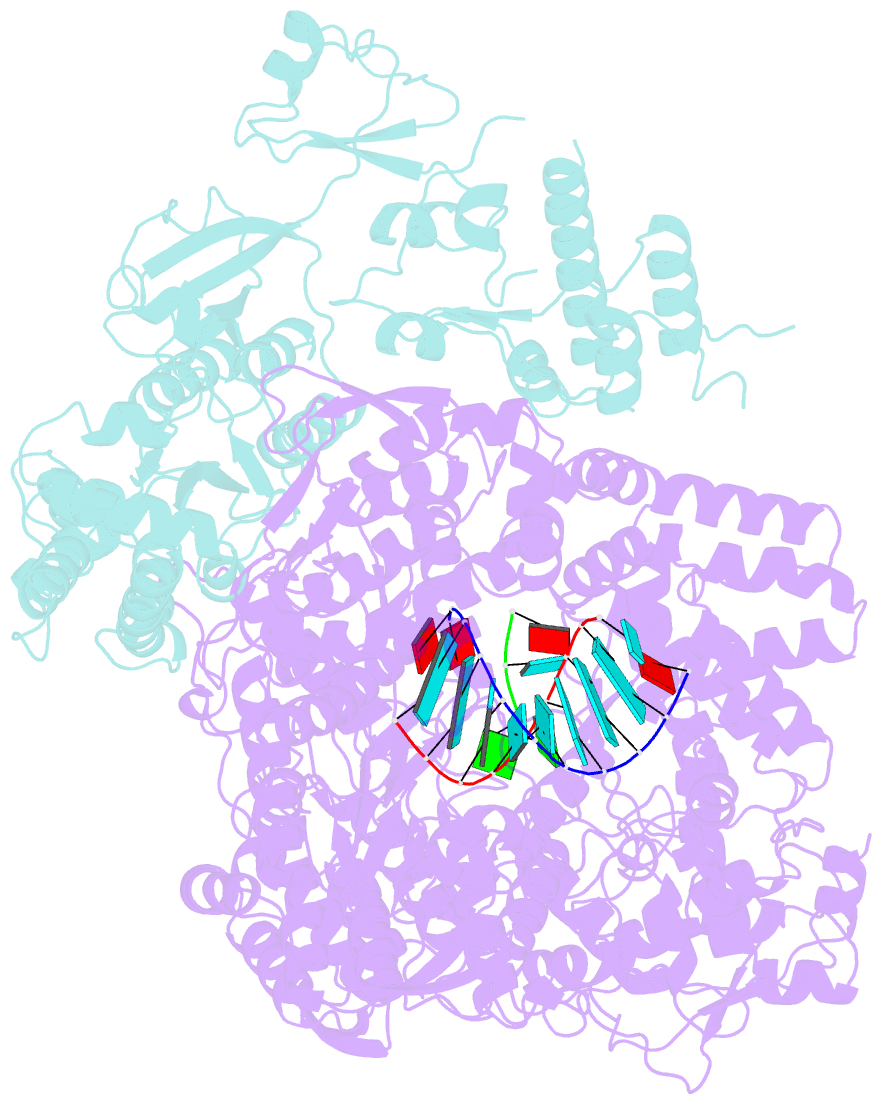
- In situ structure of bmcpv RNA-dependent RNA polymerase at early-elongation state
- Reference
- Cui Y, Zhang Y, Zhou K, Sun J, Zhou ZH (2019): "Conservative transcription in three steps visualized in a double-stranded RNA virus." Nat.Struct.Mol.Biol., 26, 1023-1034. doi: 10.1038/s41594-019-0320-0.
- Abstract
- Endogenous RNA transcription characterizes double-stranded RNA (dsRNA) viruses in the Reoviridae, a family that is exemplified by its simple, single-shelled member cytoplasmic polyhedrosis virus (CPV). Because of the lack of in situ structures of the intermediate stages of RNA-dependent RNA polymerase (RdRp) during transcription, it is poorly understood how RdRp detects environmental cues and internal transcriptional states to initiate and coordinate repeated cycles of transcript production inside the capsid. Here, we captured five high-resolution (2.8-3.5 Å) RdRp-RNA in situ structures-representing quiescent, initiation, early elongation, elongation and abortive states-under seven experimental conditions of CPV. We observed the 'Y'-form initial RNA fork in the initiation state and the complete transcription bubble in the elongation state. These structures reveal that de novo RNA transcription involves three major conformational changes during state transitions. Our results support an ouroboros model for endogenous conservative transcription in dsRNA viruses.