Summary information and primary citation
- PDB-id
- 6u7t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.0 Å)
- Summary
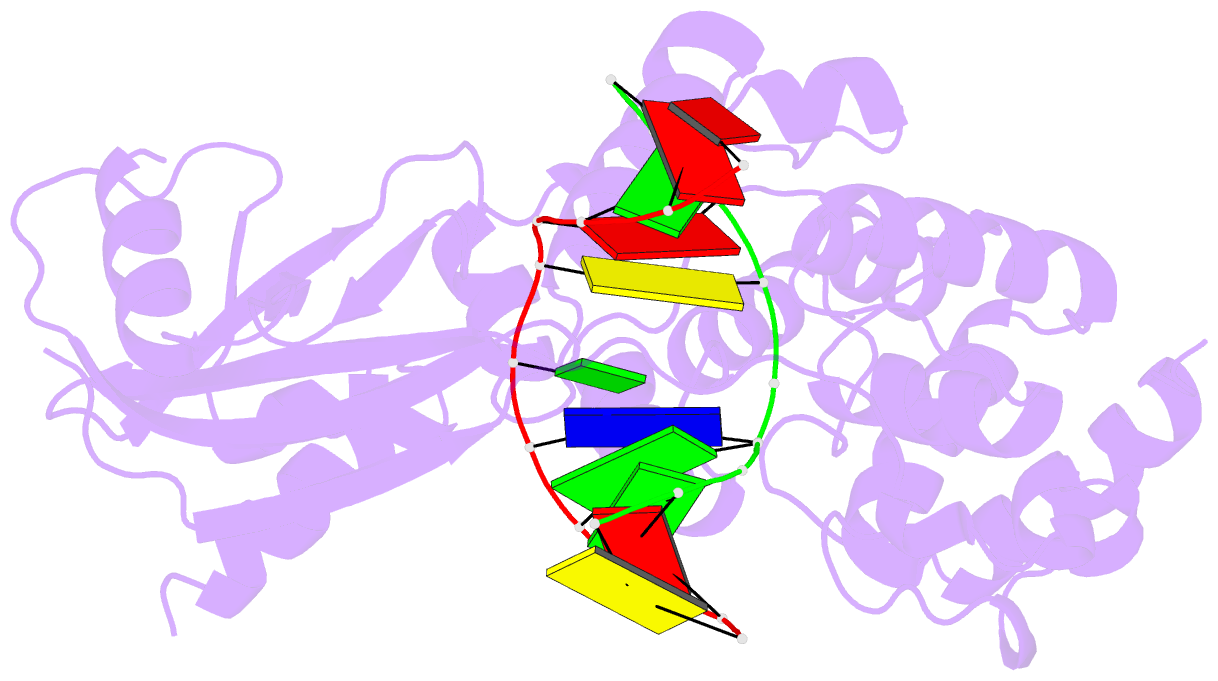
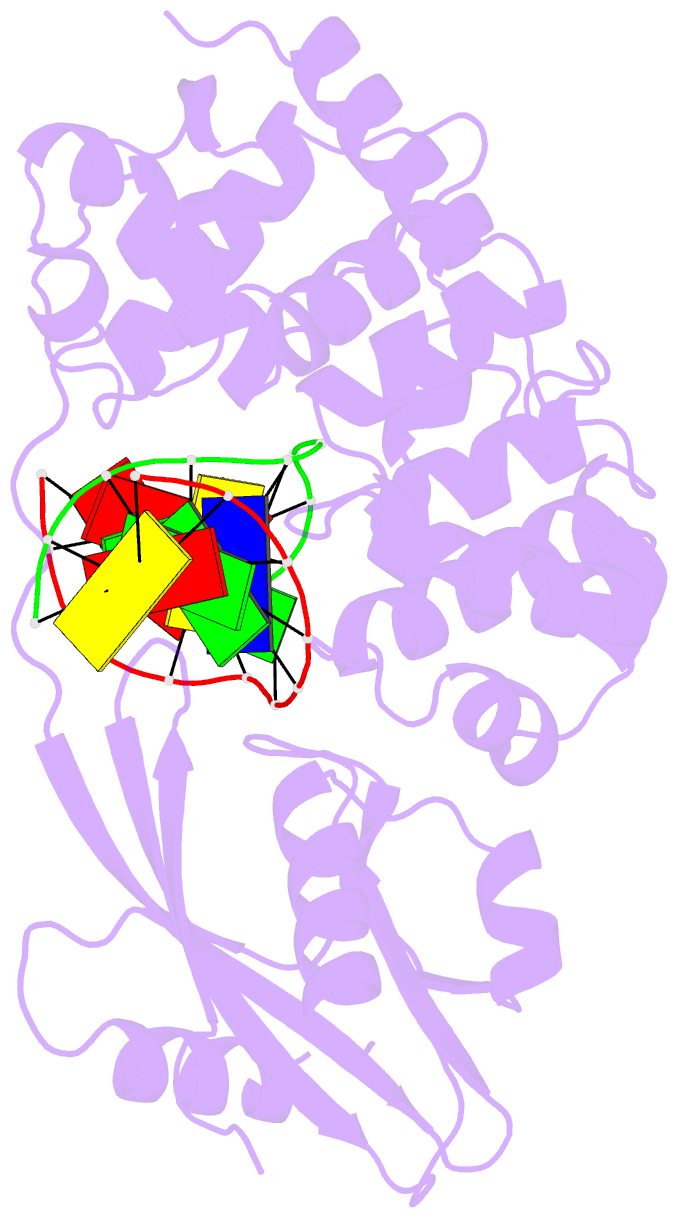
- Muty adenine glycosylase bound to DNA containing a transition state analog (1n) paired with d(8-oxo-g)
- Reference
- Russelburg LP, O'Shea Murray VL, Demir M, Knutsen KR, Sehgal SL, Cao S, David SS, Horvath MP (2020): "Structural Basis for Finding OG Lesions and Avoiding Undamaged G by the DNA Glycosylase MutY." Acs Chem.Biol., 15, 93-102. doi: 10.1021/acschembio.9b00639.
- Abstract
- The adenine glycosylase MutY selectively initiates repair of OG:A lesions and, by comparison, avoids G:A mispairs. The ability to distinguish these closely related substrates relies on the C-terminal domain of MutY, which structurally resembles MutT. To understand the mechanism for substrate specificity, we crystallized MutY in complex with DNA containing G across from the high-affinity azaribose transition state analogue. Our structure shows that G is accommodated by the OG site and highlights the role of a serine residue in OG versus G discrimination. The functional significance of Ser308 and its neighboring residues was evaluated by mutational analysis, revealing the critical importance of a β loop in the C-terminal domain for mutation suppression in cells, and biochemical performance in vitro. This loop comprising residues Phe307, Ser308, and His309 (Geobacillus stearothermophilus sequence positions) is conserved in MutY but absent in MutT and other DNA repair enzymes and may therefore serve as a MutY-specific target exploitable by chemical biological probes.