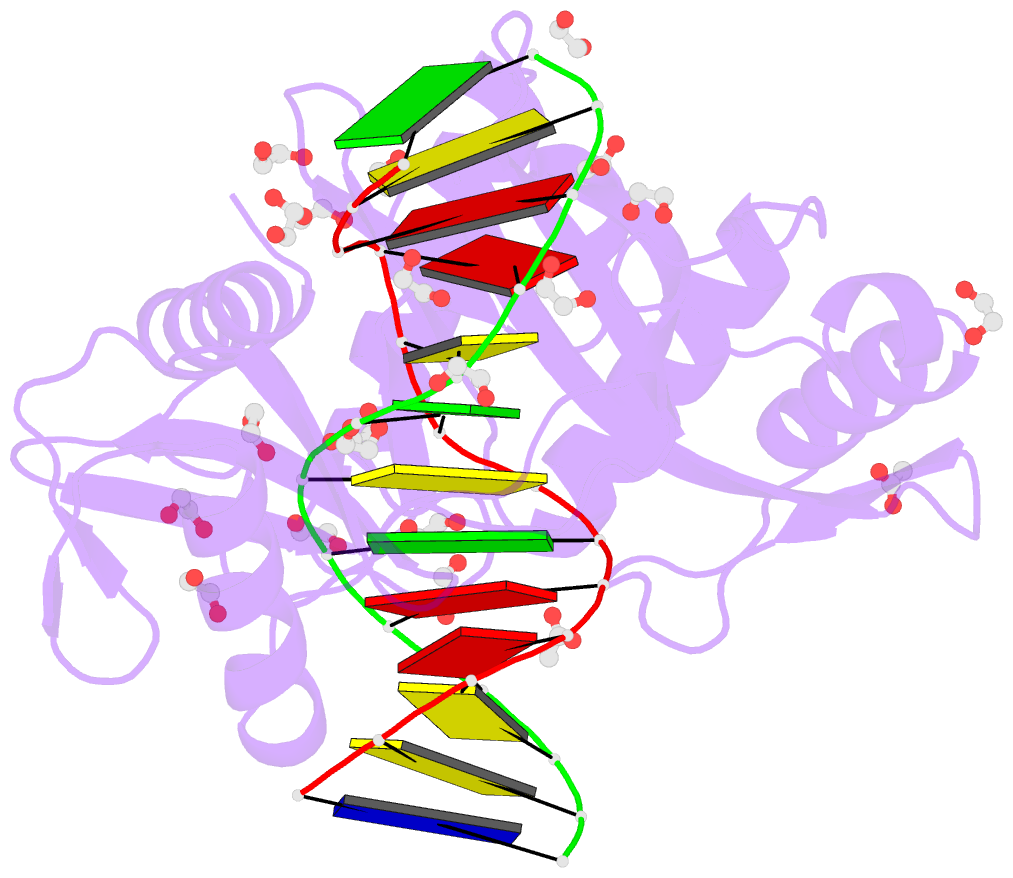
Summary information and primary citation
- PDB-id
- 6uke; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.62 Å)
- Summary
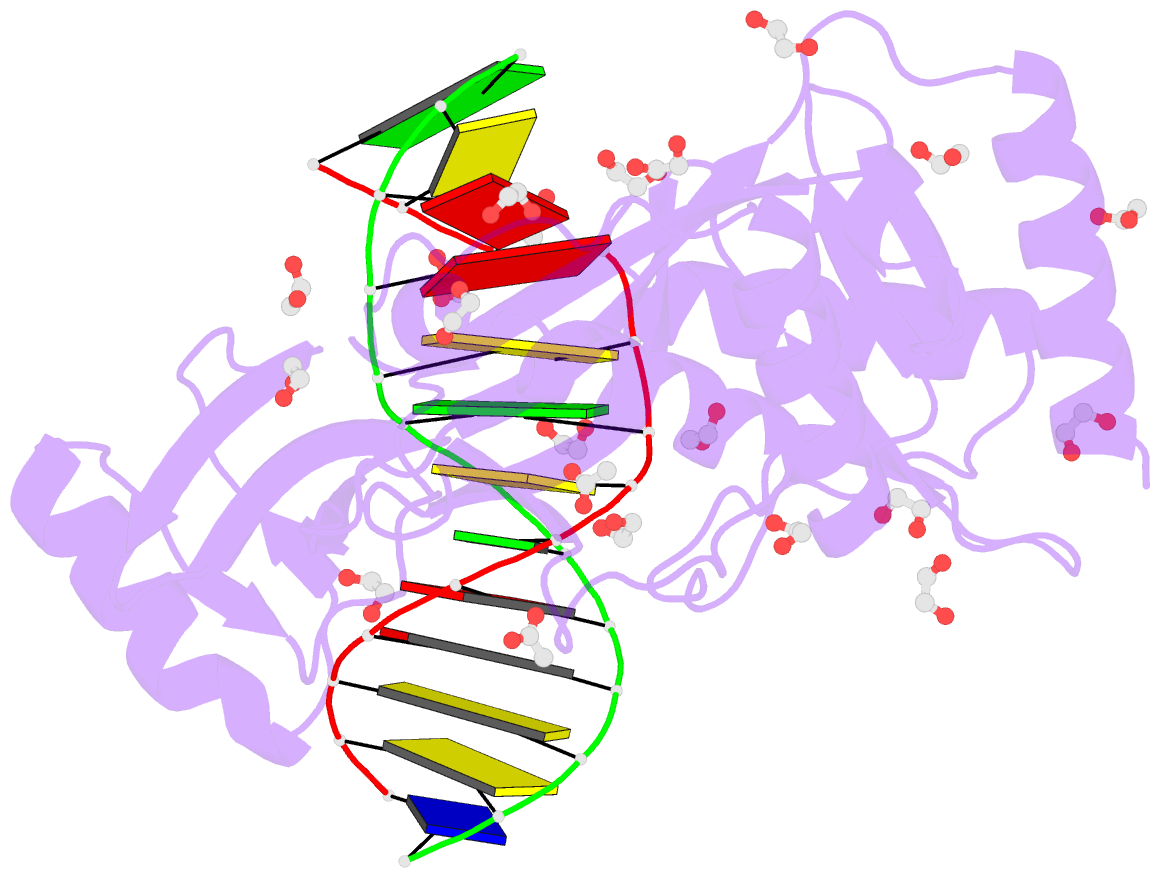
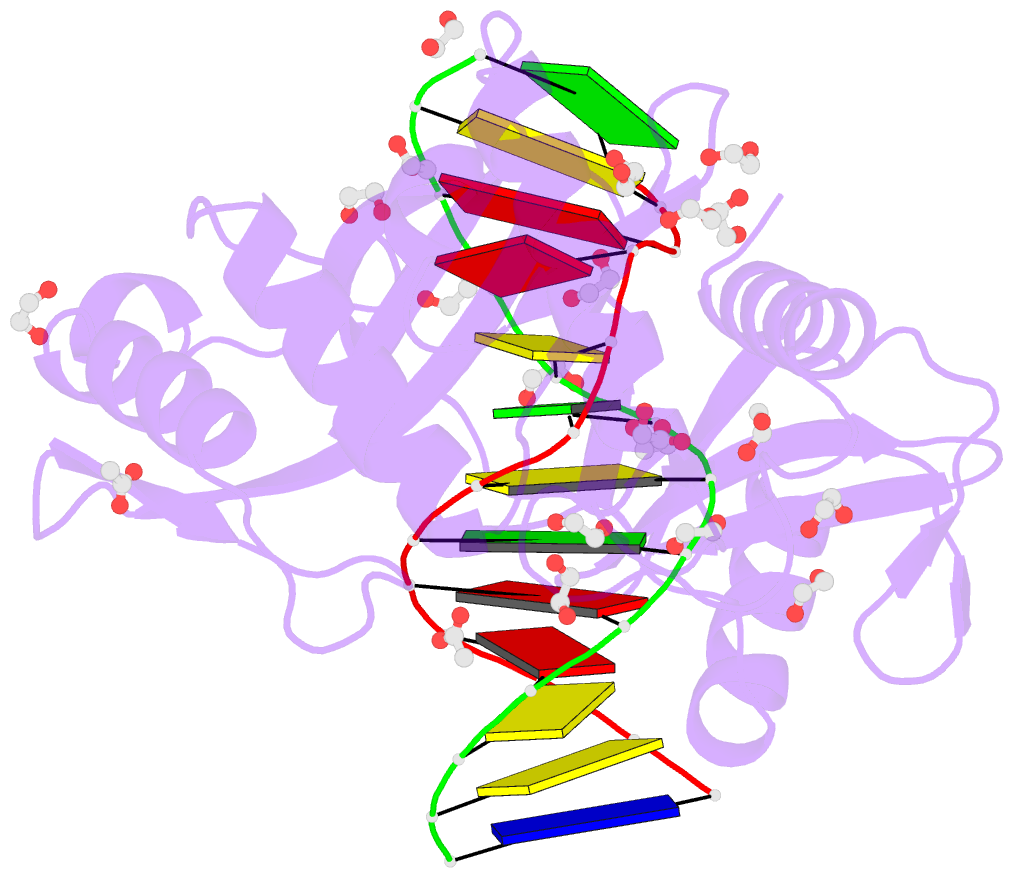
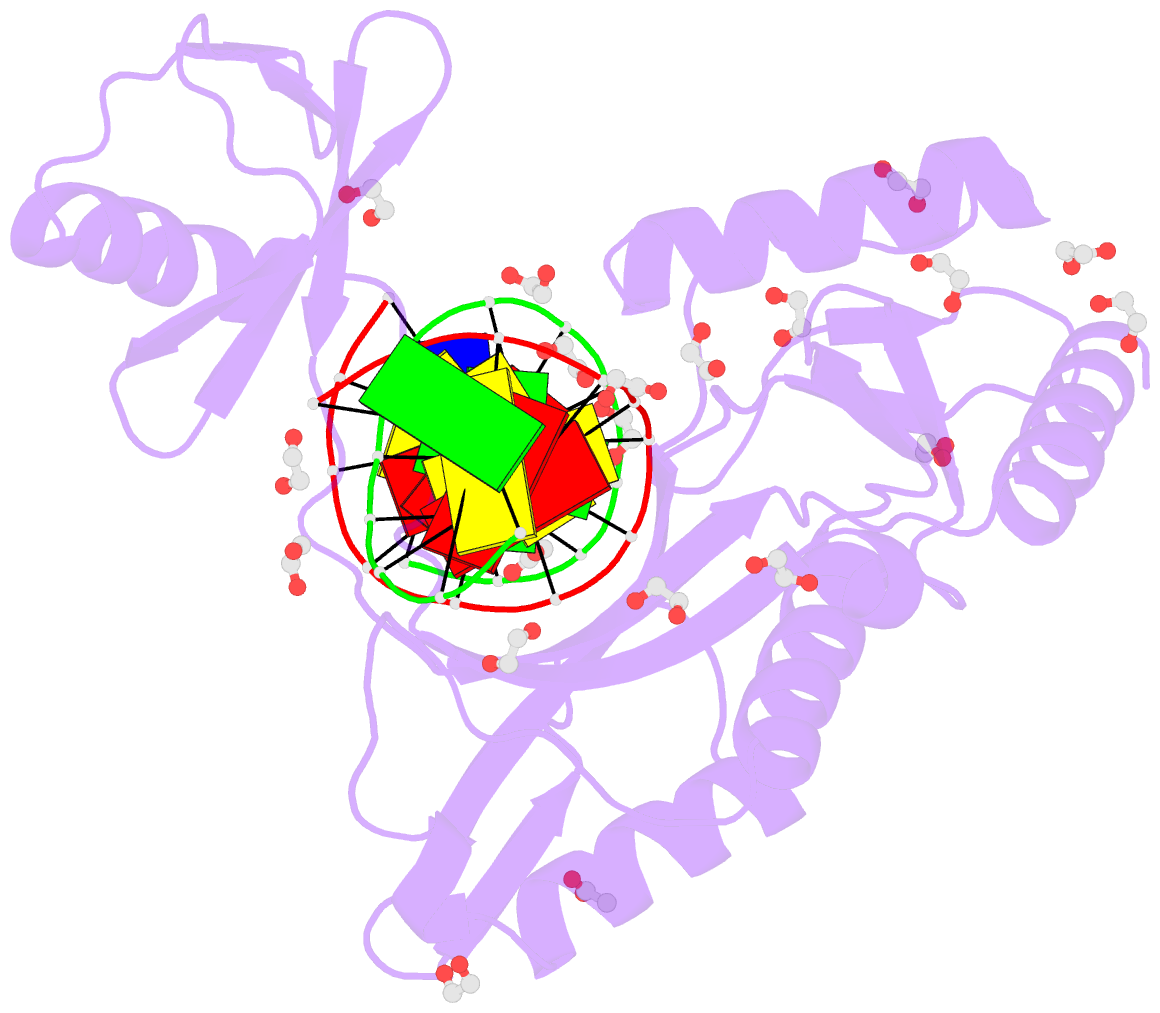
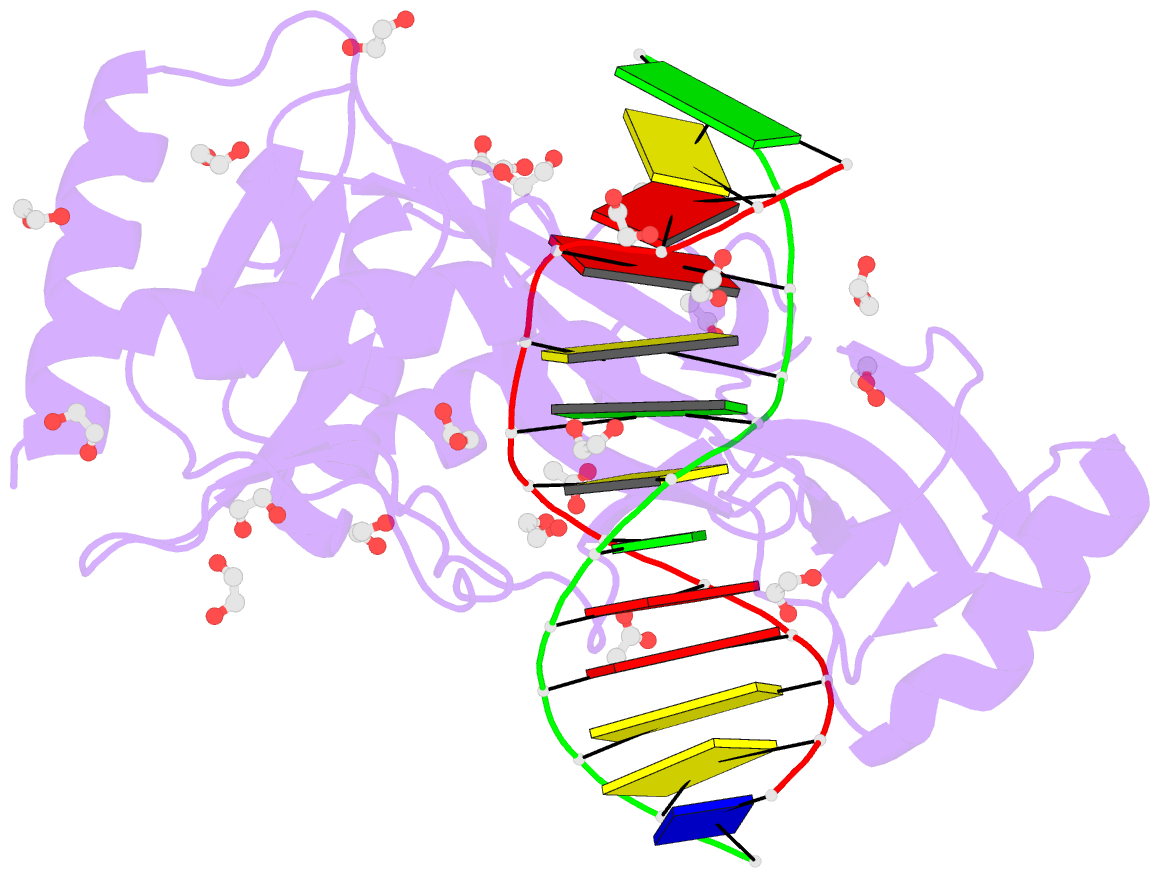
- Hhai endonuclease in complex with iodine-labelled DNA
- Reference
- Horton JR, Yang J, Zhang X, Petronzio T, Fomenkov A, Wilson GG, Roberts RJ, Cheng X (2020): "Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom." Nucleic Acids Res., 48, 1466-1478. doi: 10.1093/nar/gkz1195.
- Abstract
- HhaI, a Type II restriction endonuclease, recognizes the symmetric sequence 5'-GCG↓C-3' in duplex DNA and cleaves ('↓') to produce fragments with 2-base, 3'-overhangs. We determined the structure of HhaI in complex with cognate DNA at an ultra-high atomic resolution of 1.0 Å. Most restriction enzymes act as dimers with two catalytic sites, and cleave the two strands of duplex DNA simultaneously, in a single binding event. HhaI, in contrast, acts as a monomer with only one catalytic site, and cleaves the DNA strands sequentially, one after the other. HhaI comprises three domains, each consisting of a mixed five-stranded β sheet with a defined function. The first domain contains the catalytic-site; the second contains residues for sequence recognition; and the third contributes to non-specific DNA binding. The active-site belongs to the 'PD-D/EXK' superfamily of nucleases and contains the motif SD-X11-EAK. The first two domains are similar in structure to two other monomeric restriction enzymes, HinP1I (G↓CGC) and MspI (C↓CGG), which produce fragments with 5'-overhangs. The third domain, present only in HhaI, shifts the positions of the recognition residues relative to the catalytic site enabling this enzyme to cleave the recognition sequence at a different position. The structure of M.HhaI, the biological methyltransferase partner of HhaI, was determined earlier. Together, these two structures represent the first natural pair of restriction-modification enzymes to be characterized in atomic detail.