Summary information and primary citation
- PDB-id
- 6upk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (4.9 Å)
- Summary
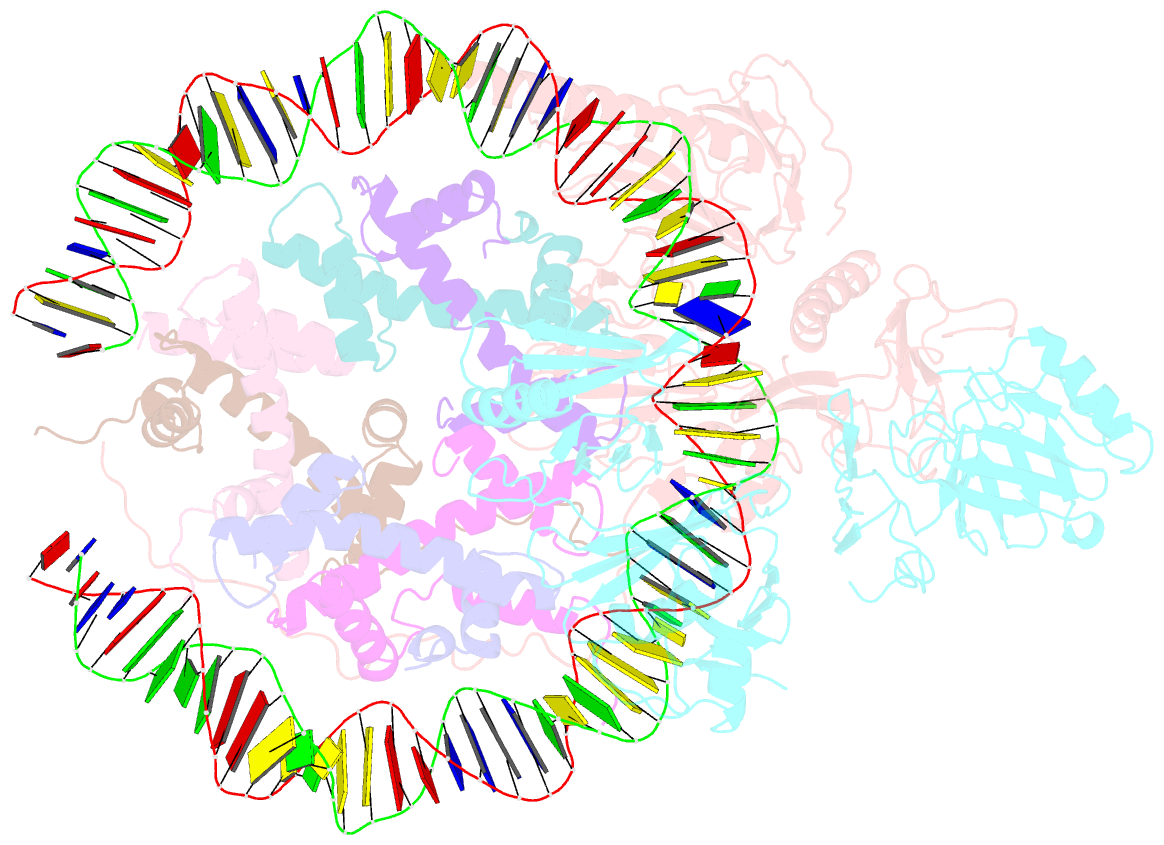
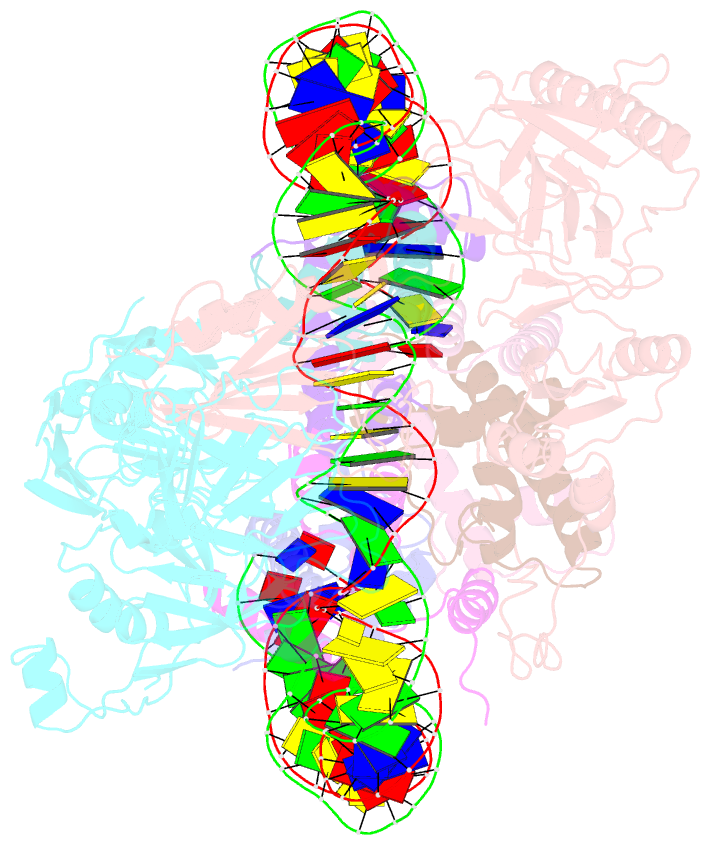
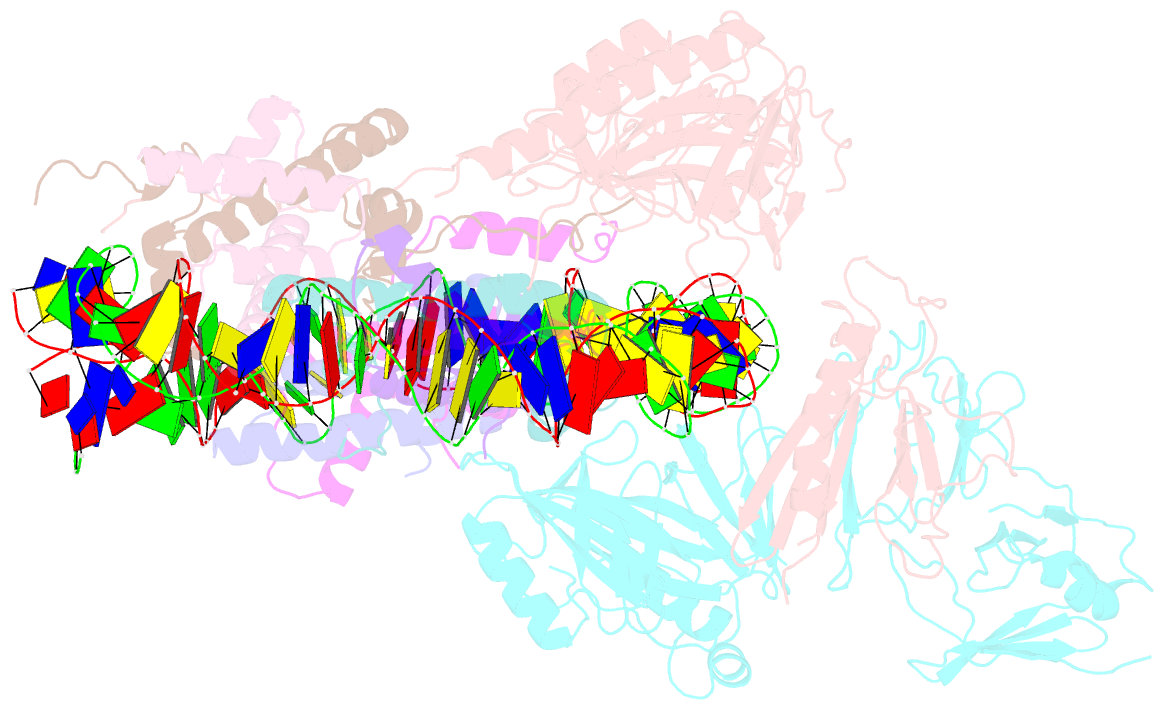
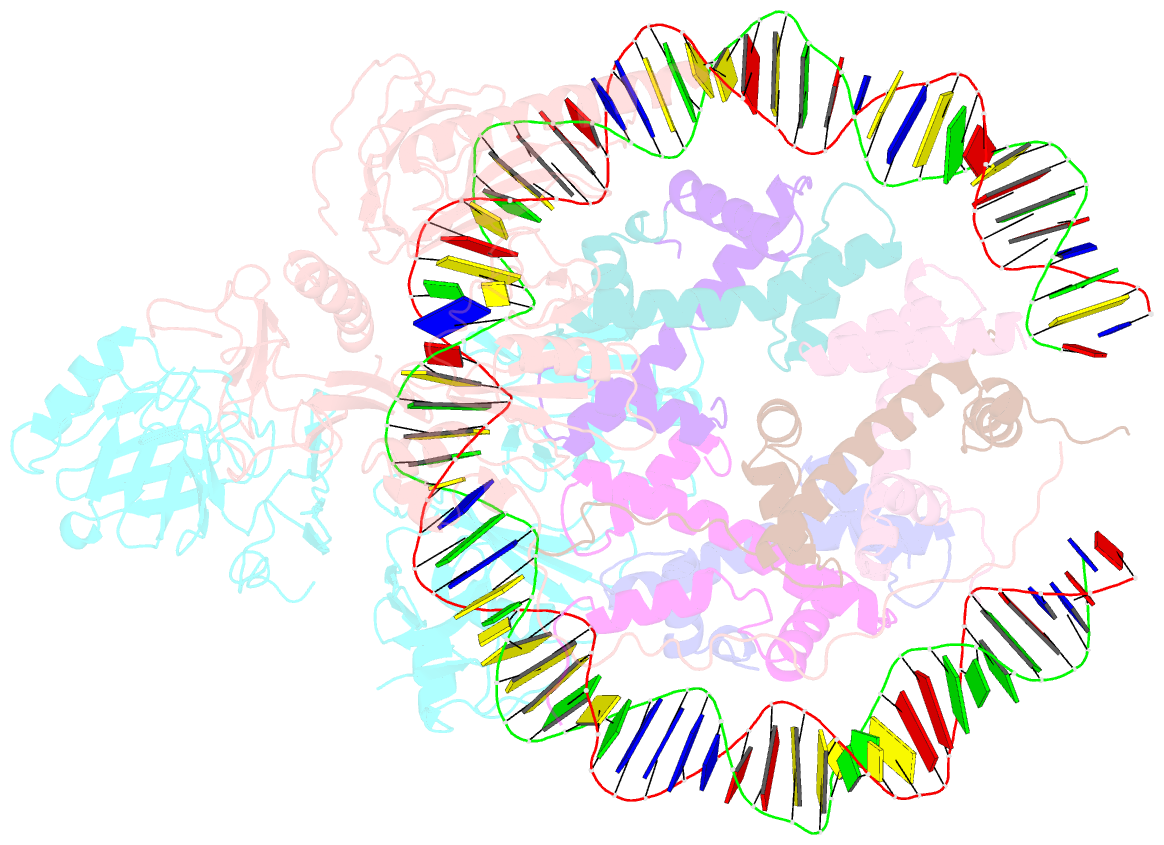
- Structure of fact_subnucleosome complex 1
- Reference
- Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D'Arcy S, Luger K (2020): "FACT caught in the act of manipulating the nucleosome." Nature, 577, 426-431. doi: 10.1038/s41586-019-1820-0.
- Abstract
- The organization of genomic DNA into nucleosomes profoundly affects all DNA-related processes in eukaryotes. The histone chaperone known as 'facilitates chromatin transcription' (FACT1) (consisting of subunits SPT16 and SSRP1) promotes both disassembly and reassembly of nucleosomes during gene transcription, DNA replication and DNA repair2. However, the mechanism by which FACT causes these opposing outcomes is unknown. Here we report two cryo-electron-microscopic structures of human FACT in complex with partially assembled subnucleosomes, with supporting biochemical and hydrogen-deuterium exchange data. We find that FACT is engaged in extensive interactions with nucleosomal DNA and all histone variants. The large DNA-binding surface on FACT appears to be protected by the carboxy-terminal domains of both of its subunits, and this inhibition is released by interaction with H2A-H2B, allowing FACT-H2A-H2B to dock onto a complex containing DNA and histones H3 and H4 (ref. 3). SPT16 binds nucleosomal DNA and tethers H2A-H2B through its carboxy-terminal domain by acting as a placeholder for DNA. SSRP1 also contributes to DNA binding, and can assume two conformations, depending on whether a second H2A-H2B dimer is present. Our data suggest a compelling mechanism for how FACT maintains chromatin integrity during polymerase passage, by facilitating removal of the H2A-H2B dimer, stabilizing intermediate subnucleosomal states and promoting nucleosome reassembly. Our findings reconcile discrepancies regarding the many roles of FACT and underscore the dynamic interactions between histone chaperones and nucleosomes.