Summary information and primary citation
- PDB-id
- 6v4x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (3.2 Å)
- Summary
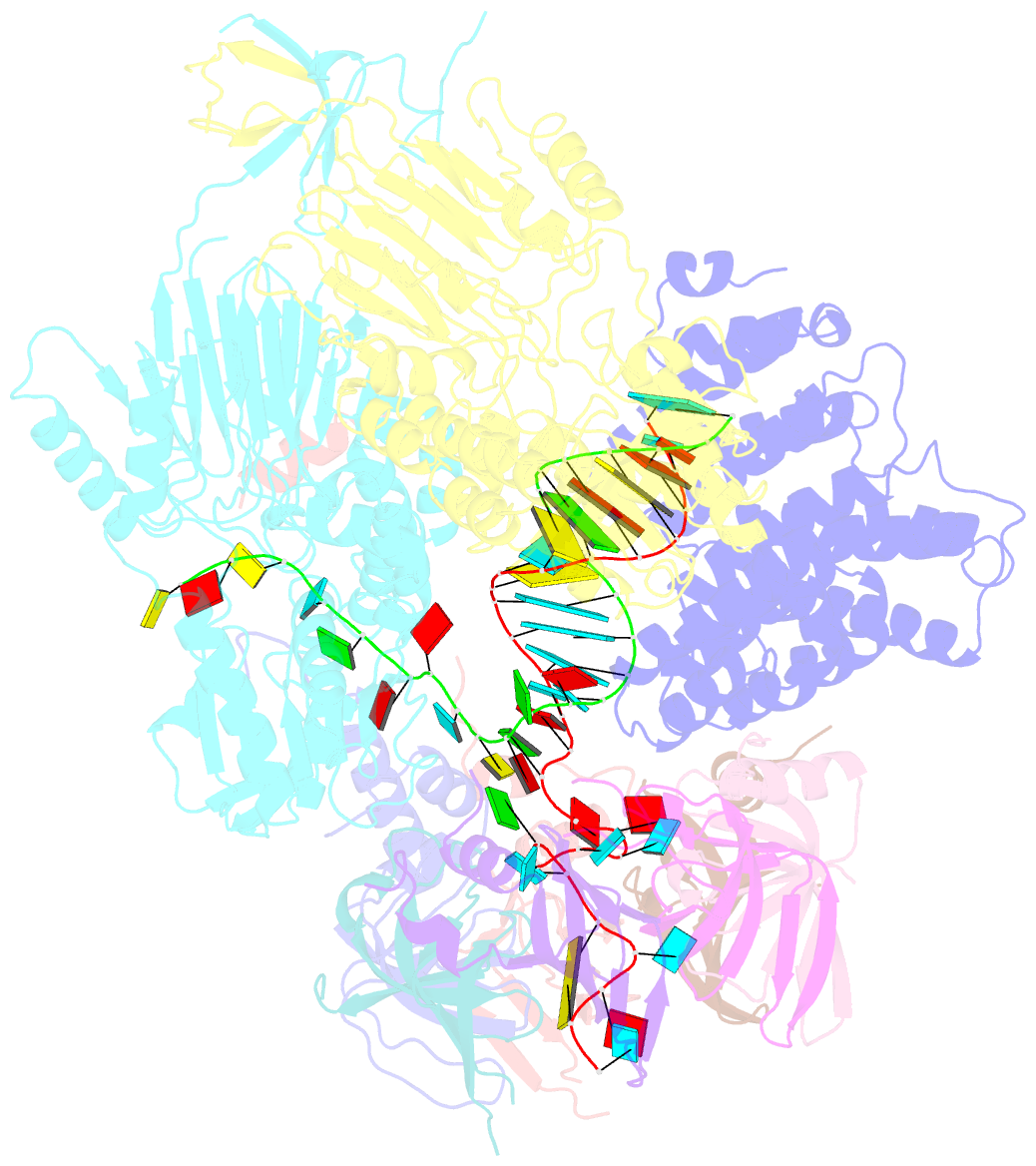
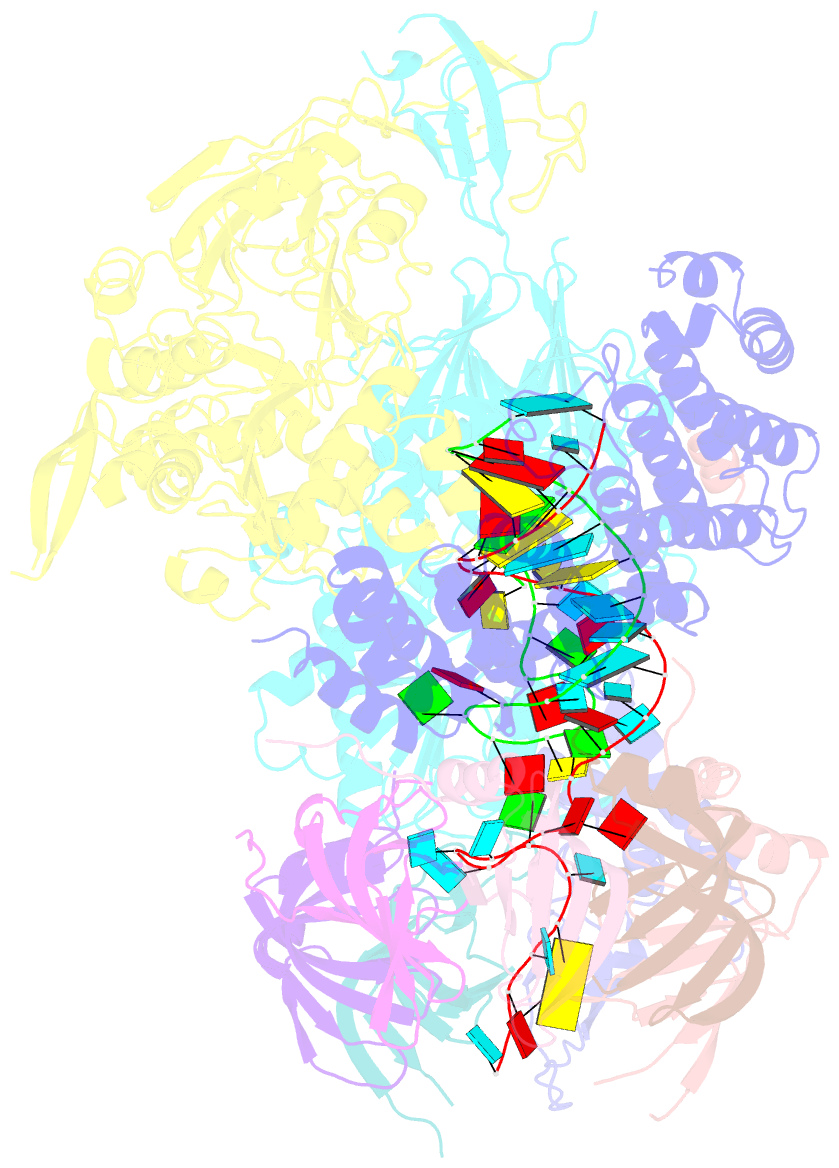
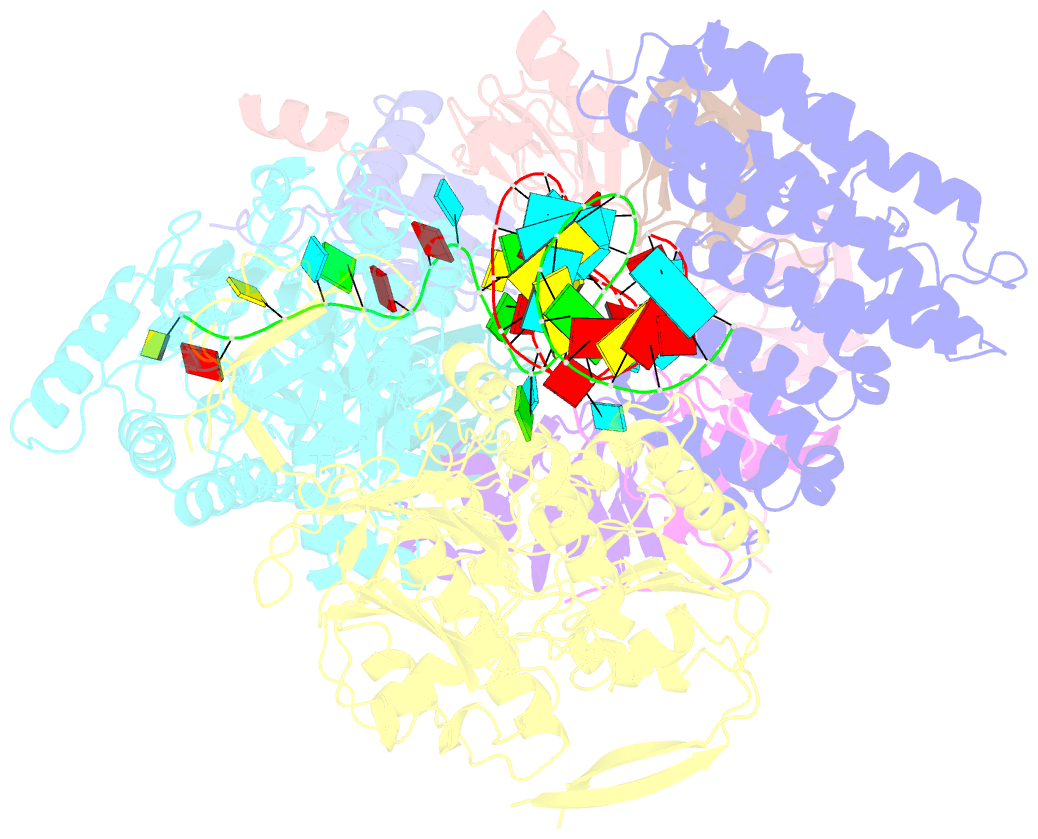
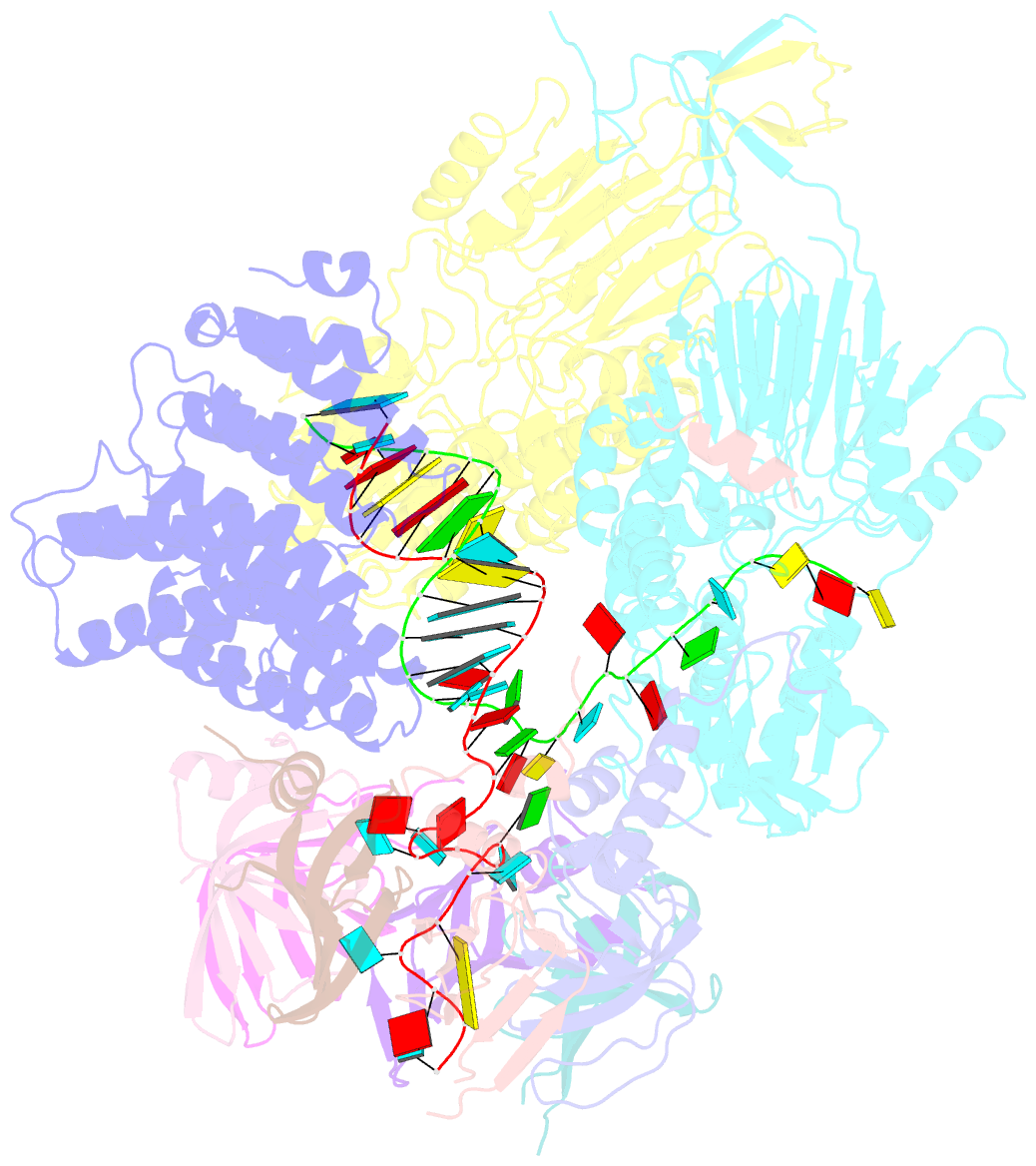
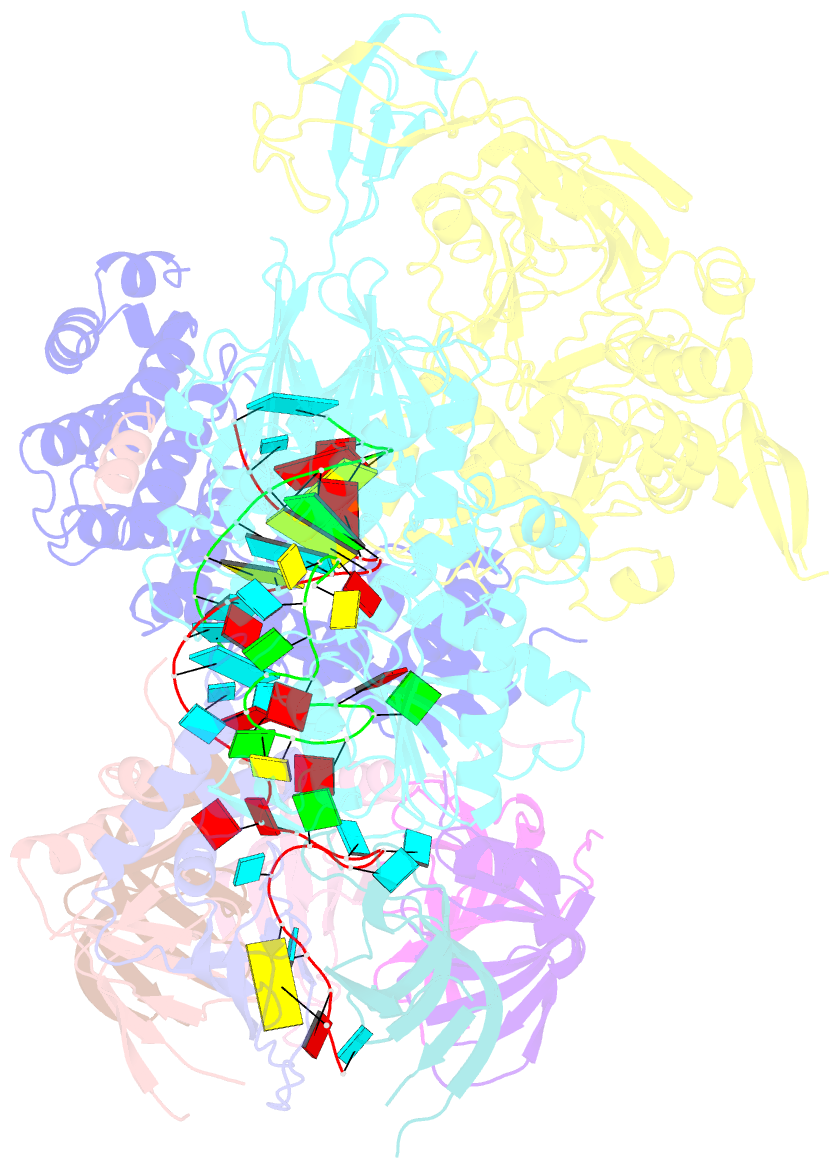
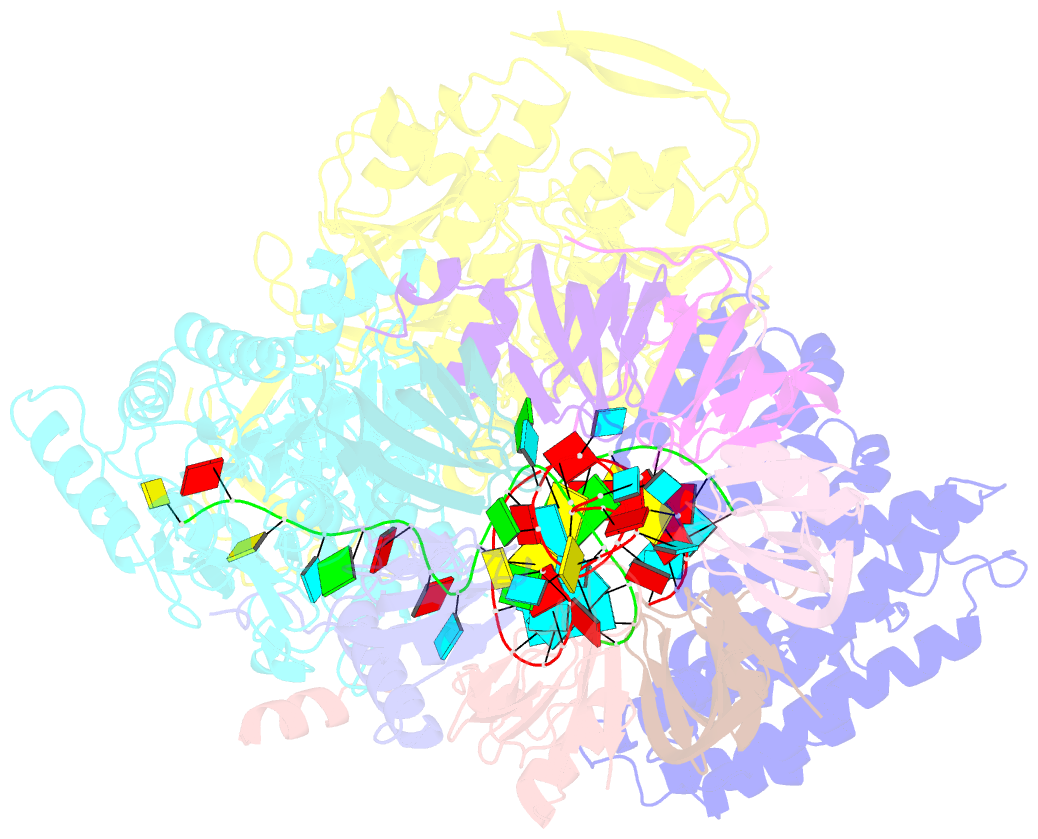
- cryo-EM structure of an active human histone pre-mrna 3'-end processing machinery at 3.2 angstrom resolution
- Reference
- Sun Y, Zhang Y, Aik WS, Yang XC, Marzluff WF, Walz T, Dominski Z, Tong L (2020): "Structure of an active human histone pre-mRNA 3'-end processing machinery." Science, 367, 700-703. doi: 10.1126/science.aaz7758.
- Abstract
- The 3'-end processing machinery for metazoan replication-dependent histone precursor messenger RNAs (pre-mRNAs) contains the U7 small nuclear ribonucleoprotein and shares the key cleavage module with the canonical cleavage and polyadenylation machinery. We reconstituted an active human histone pre-mRNA processing machinery using 13 recombinant proteins and two RNAs and determined its structure by cryo-electron microscopy. The overall structure is highly asymmetrical and resembles an amphora with one long handle. We captured the pre-mRNA in the active site of the endonuclease, the 73-kilodalton subunit of the cleavage and polyadenylation specificity factor, poised for cleavage. The endonuclease and the entire cleavage module undergo extensive rearrangements for activation, triggered through the recognition of the duplex between the authentic pre-mRNA and U7 small nuclear RNA (snRNA). Our study also has notable implications for understanding canonical and snRNA 3'-end processing.