Summary information and primary citation
- PDB-id
- 6vug; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- protein binding
- Method
- X-ray (3.0 Å)
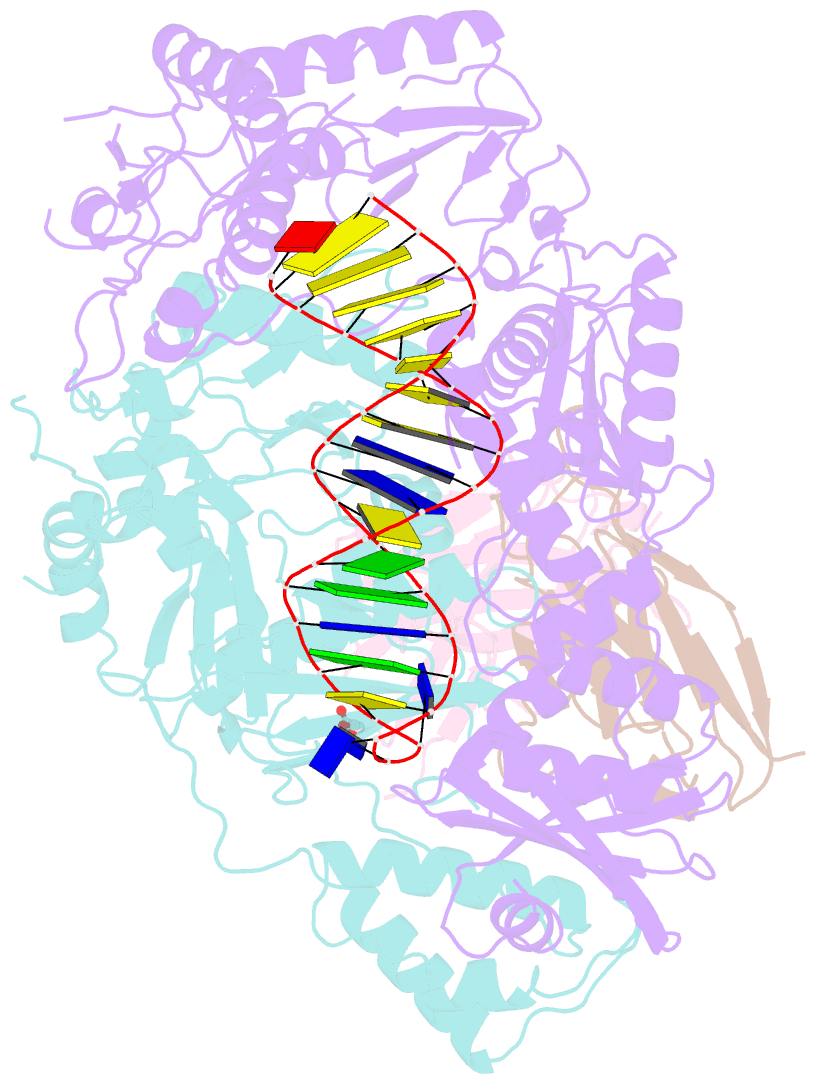
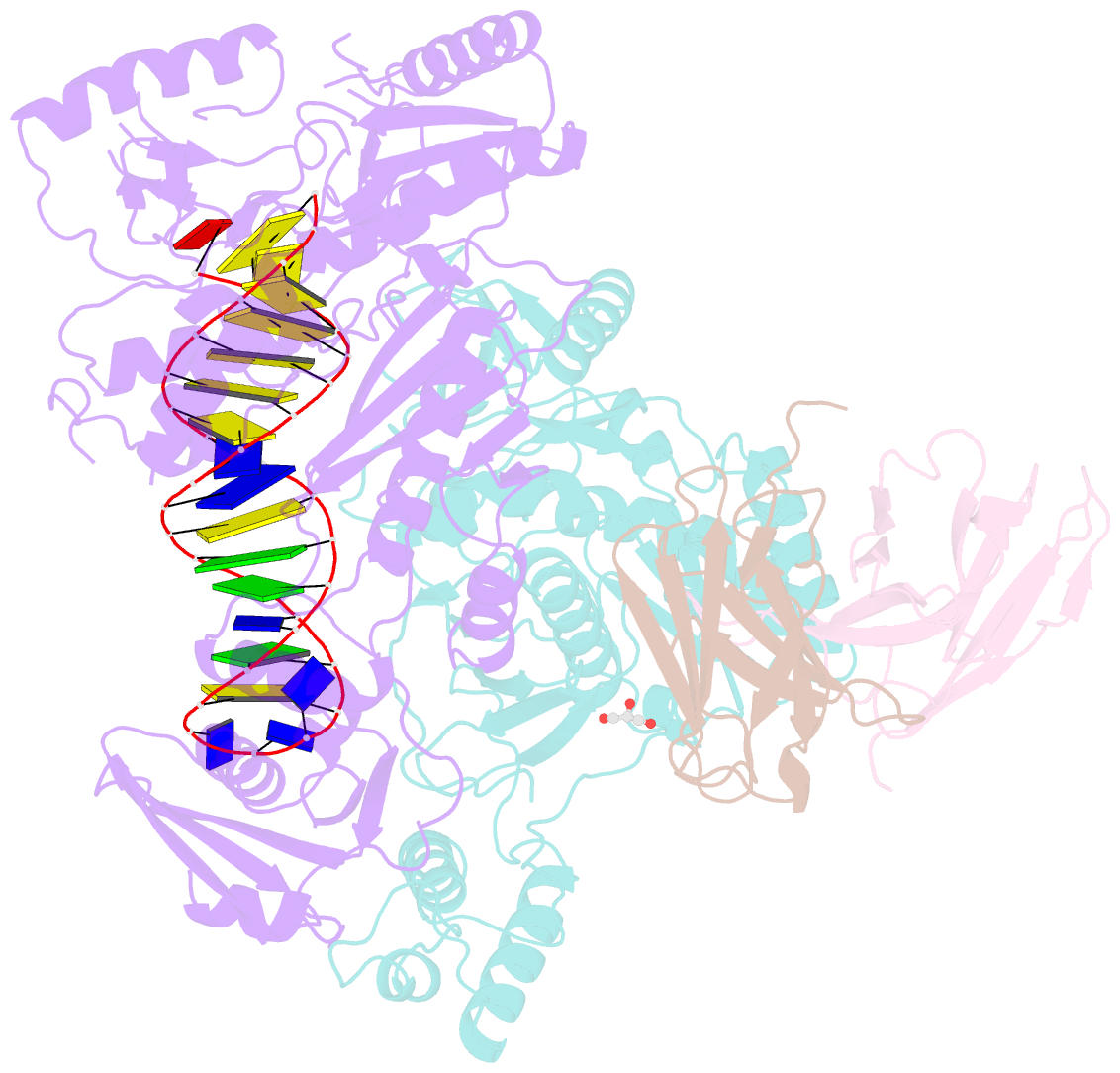
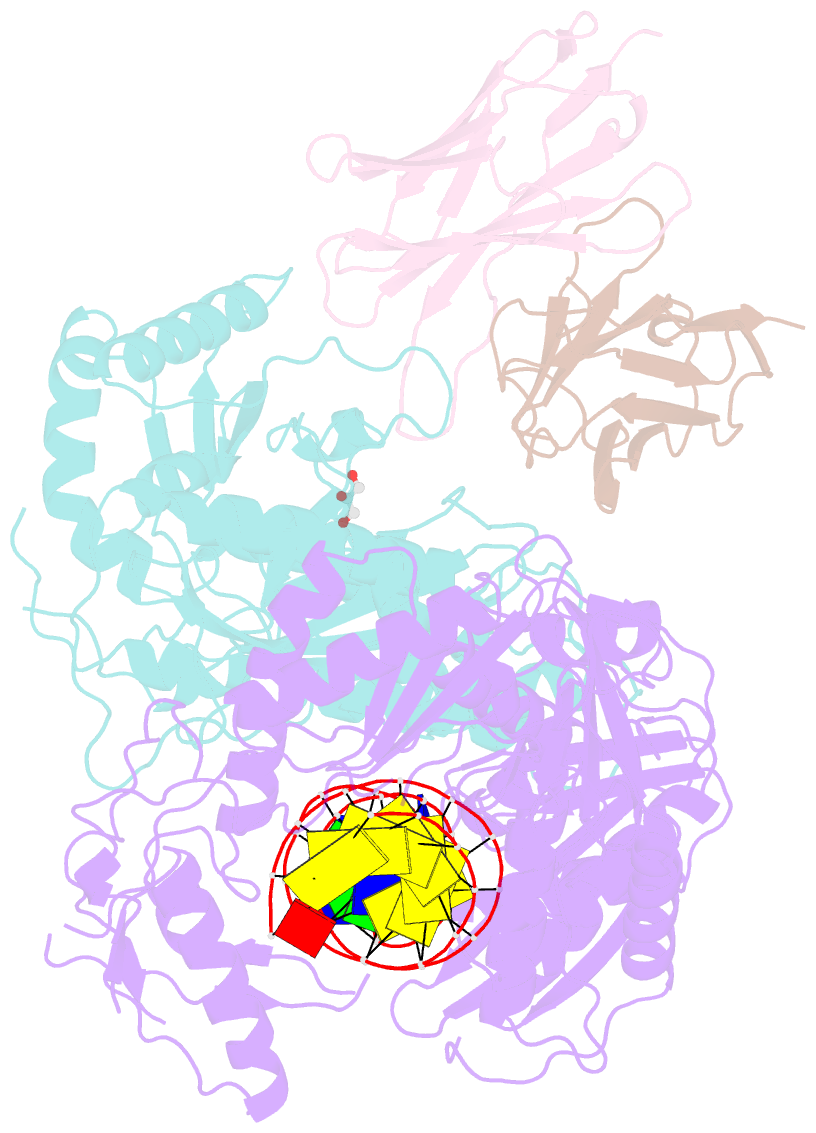
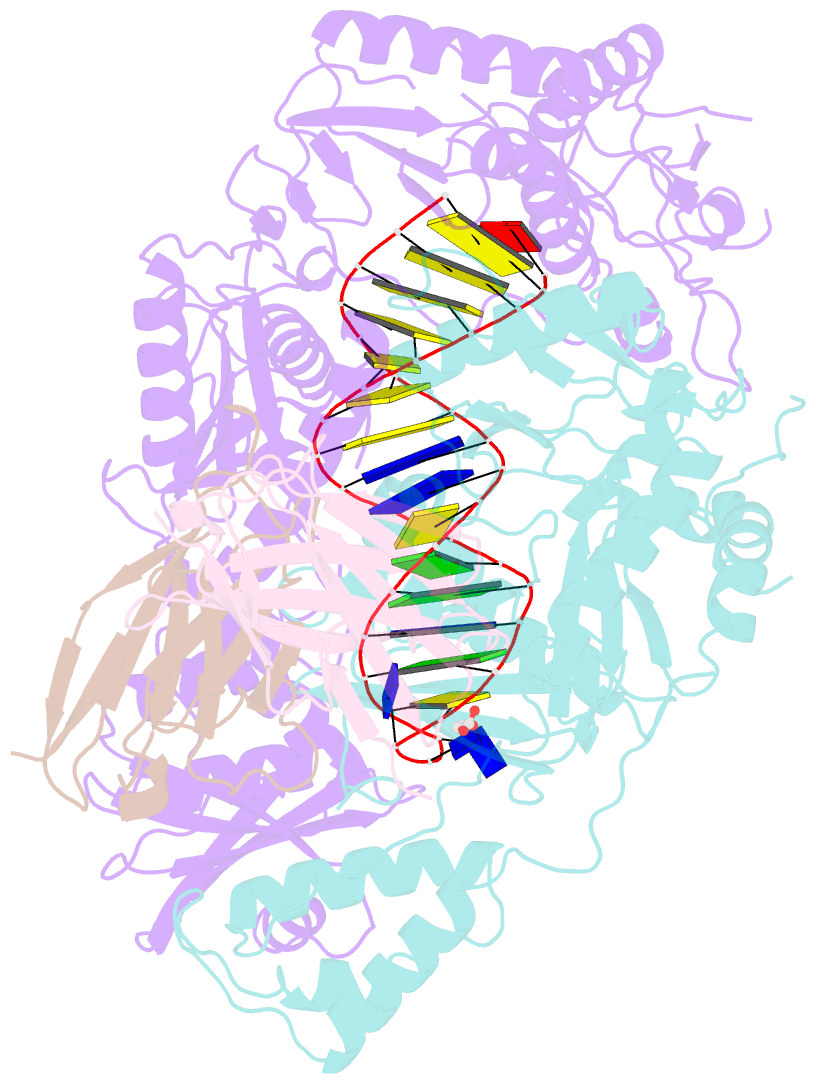
- Summary
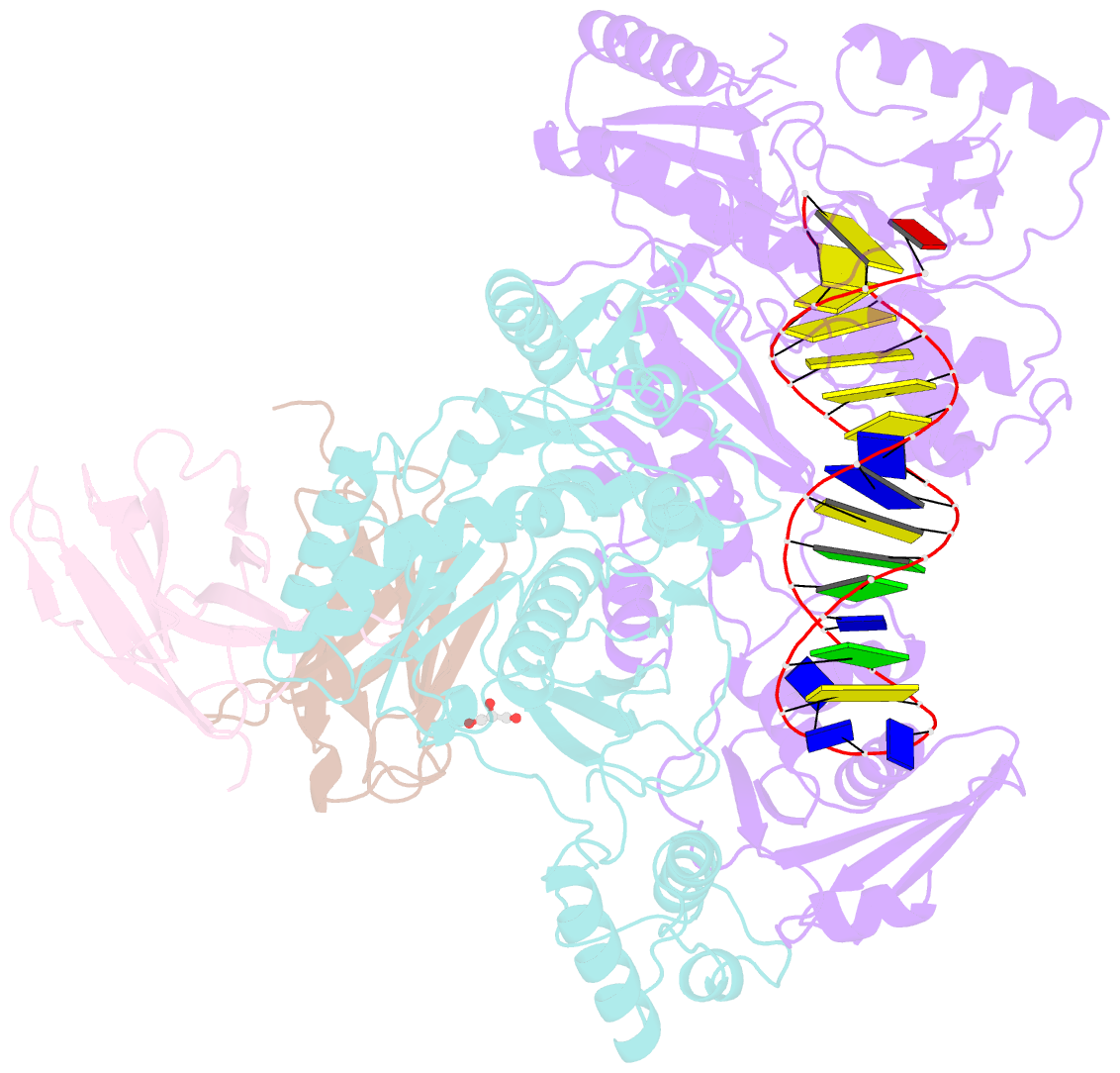
- Diabody bound to a reverse transcriptase aptamer complex
- Reference
- Chesterman C, Arnold E (2021): "Co-crystallization with diabodies: A case study for the introduction of synthetic symmetry." Structure, 29, 598-605.e3. doi: 10.1016/j.str.2021.02.001.
- Abstract
- This work presents a method for introducing synthetic symmetry into protein crystallization samples using an antibody fragment termed a diabody (Dab). These Dabs contain two target binding sites, and engineered disulfide bonds have been included to modulate Dab flexibility. The impacts of Dab engineering have been observed through assessment of thermal stability, small-angle X-ray scattering, and high-resolution crystal structures. Complexes between the engineered Dabs and HIV-1 reverse transcriptase (RT) bound to a high-affinity DNA aptamer were also generated to explore the capacity of engineered Dabs to enable the crystallization of bound target proteins. This strategy increased the crystallization hit frequency obtained for RT-aptamer, and the structure of a Dab-RT-aptamer complex was determined to 3.0-Å resolution. Introduction of synthetic symmetry using a Dab could be a broadly applicable strategy, especially when monoclonal antibodies for a target have previously been identified.