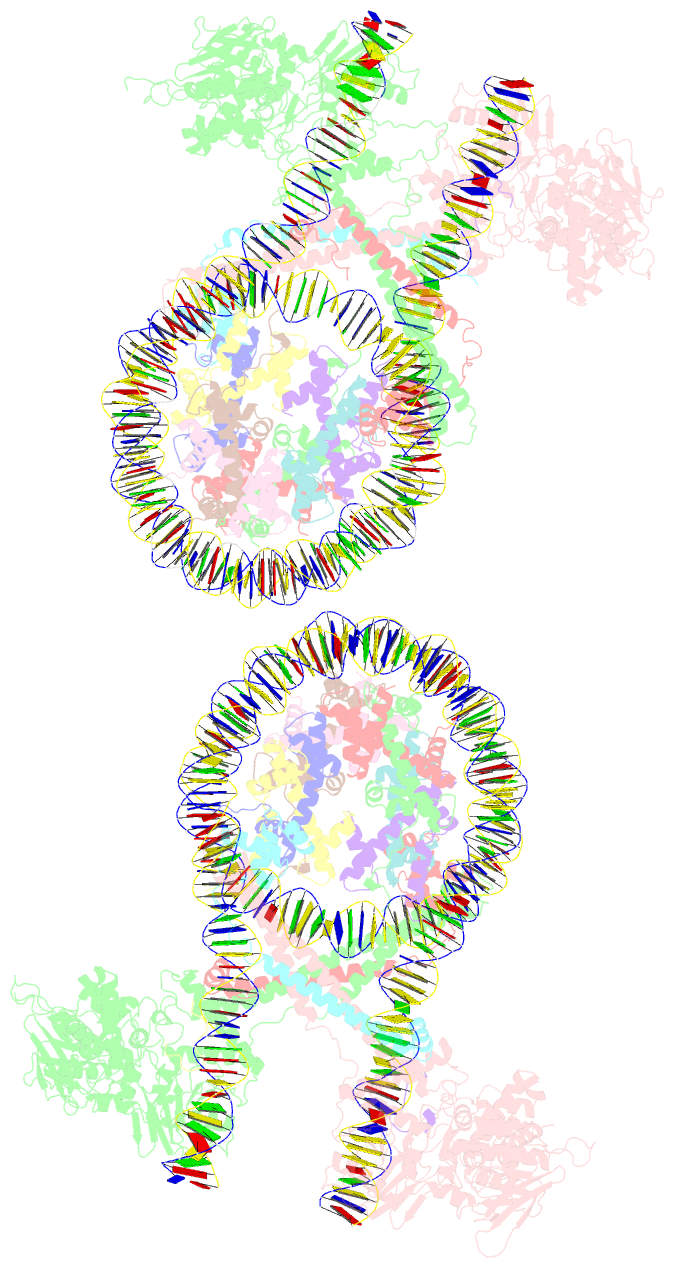
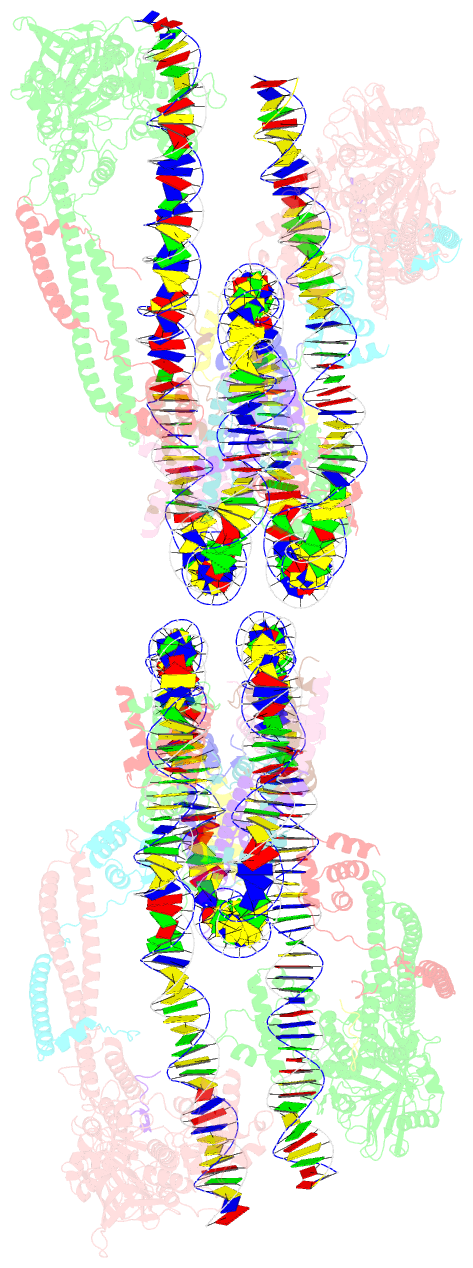
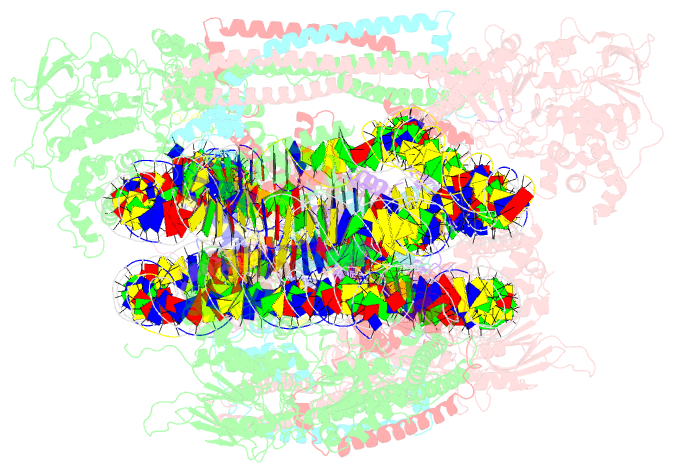
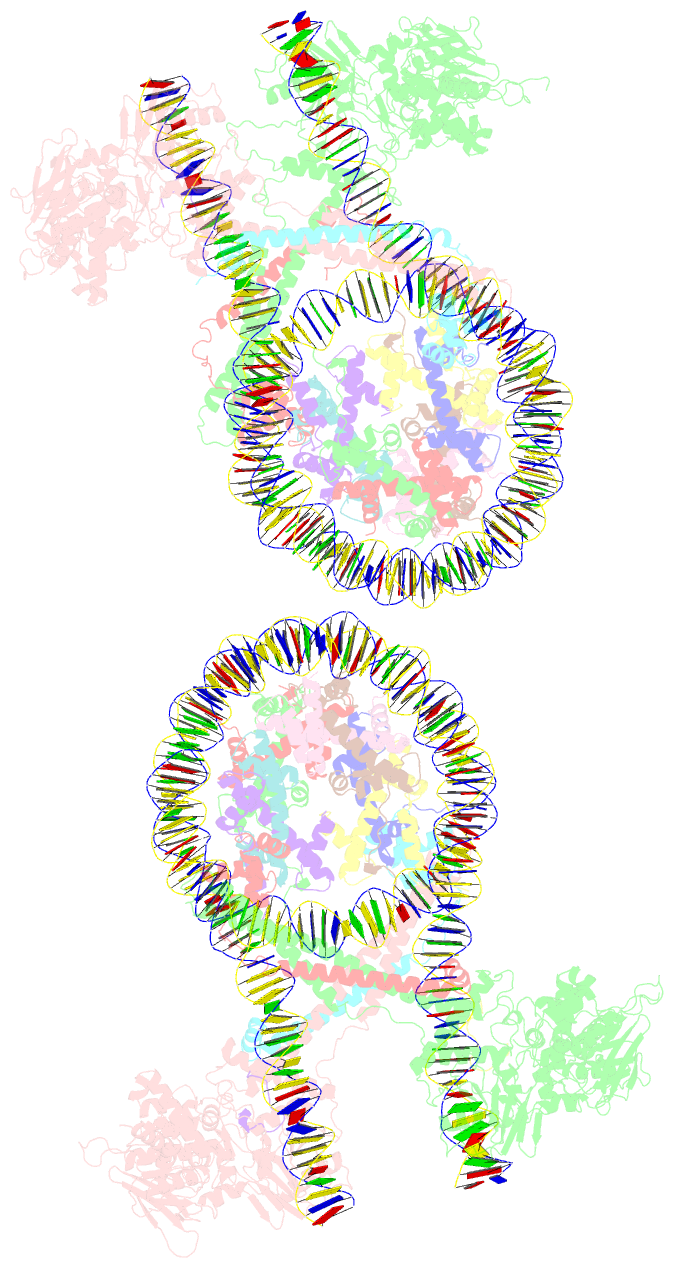
Summary information and primary citation
- PDB-id
- 6vyp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein-DNA
- Method
- X-ray (4.99 Å)
- Summary
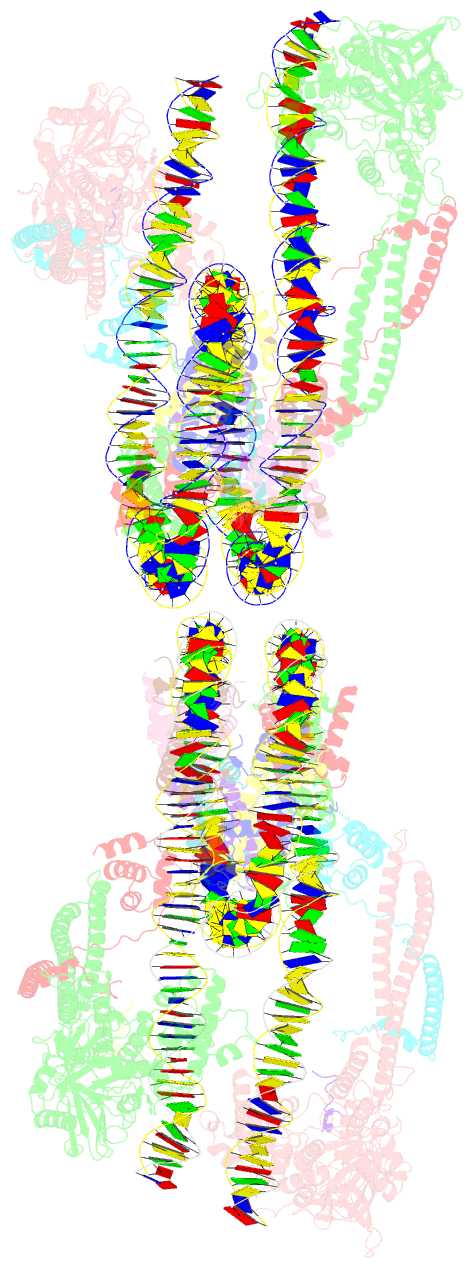
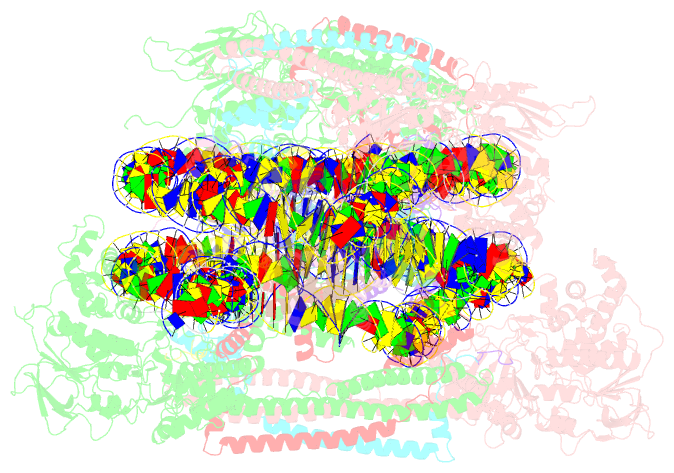
- Crystal structure of the lsd1-corest histone demethylase bound to its nucleosome substrate
- Reference
- Kim SA, Zhu J, Yennawar N, Eek P, Tan S (2020): "Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate." Mol.Cell, 78, 903. doi: 10.1016/j.molcel.2020.04.019.
- Abstract
- LSD1 (lysine specific demethylase; also known as KDM1A), the first histone demethylase discovered, regulates cell-fate determination and is overexpressed in multiple cancers. LSD1 demethylates histone H3 Lys4, an epigenetic mark for active genes, but requires the CoREST repressor to act on nucleosome substrates. To understand how an accessory subunit (CoREST) enables a chromatin enzyme (LSD1) to function on a nucleosome and not just histones, we have determined the crystal structure of the LSD1/CoREST complex bound to a 191-bp nucleosome. We find that the LSD1 catalytic domain binds extranucleosomal DNA and is unexpectedly positioned 100 Å away from the nucleosome core. CoREST makes critical contacts with both histone and DNA components of the nucleosome, explaining its essential function in demethylating nucleosome substrates. Our studies also show that the LSD1(K661A) frequently used as a catalytically inactive mutant in vivo (based on in vitro peptide studies) actually retains substantial H3K4 demethylase activity on nucleosome substrates.