Summary information and primary citation
- PDB-id
- 6vz4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- motor protein
- Method
- cryo-EM (3.9 Å)
- Summary
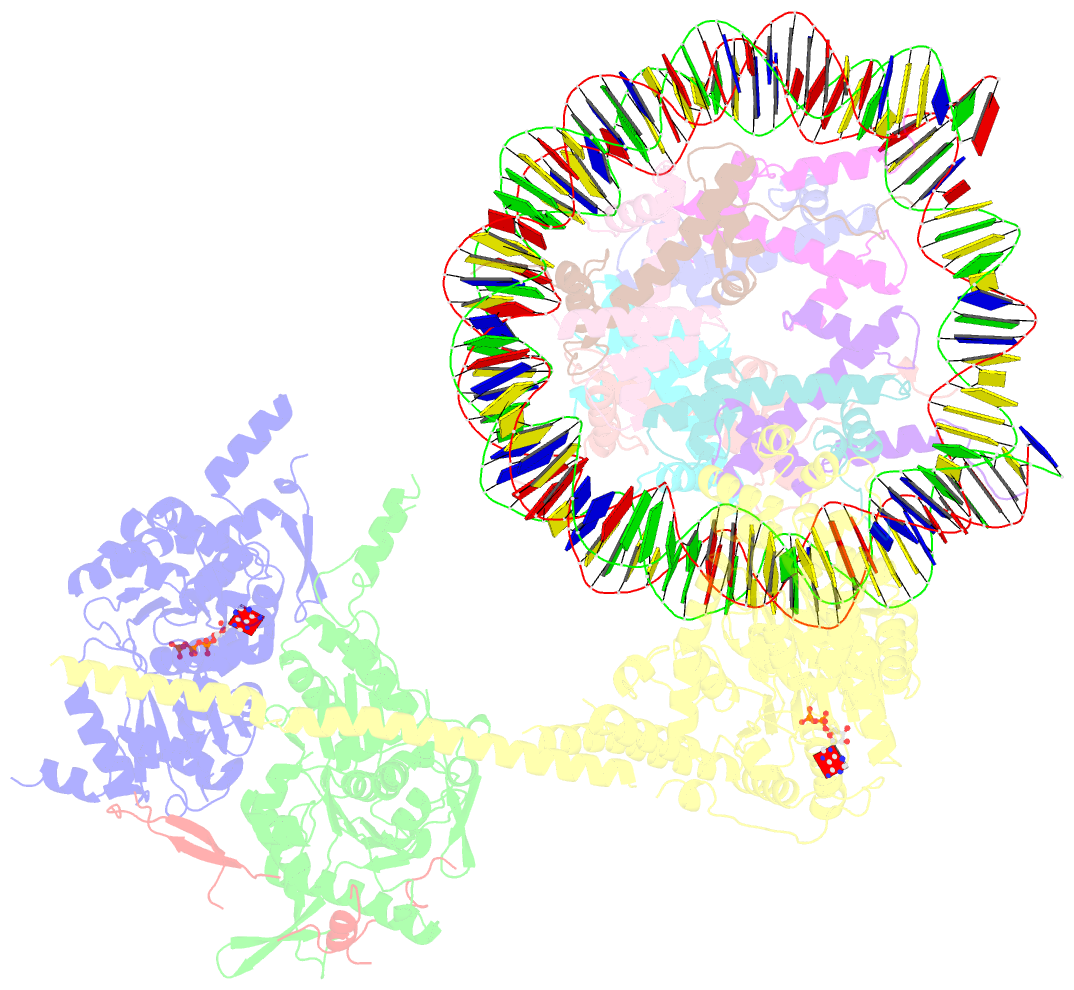
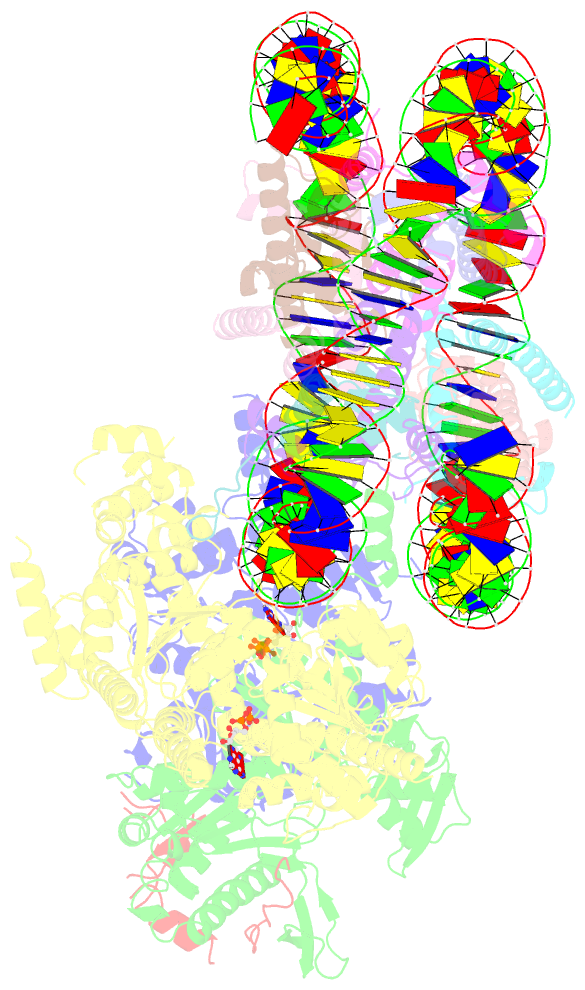
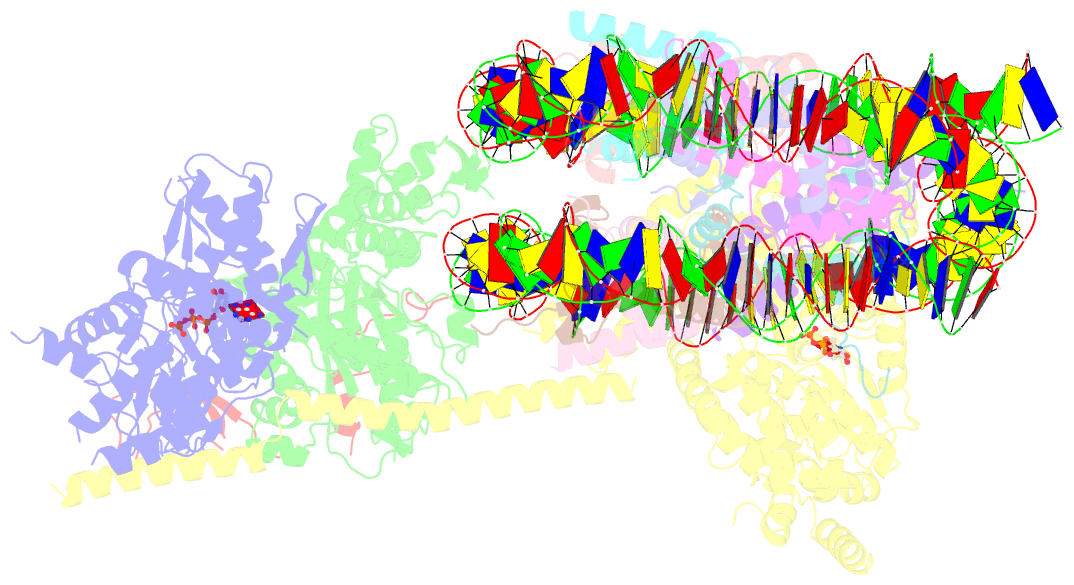
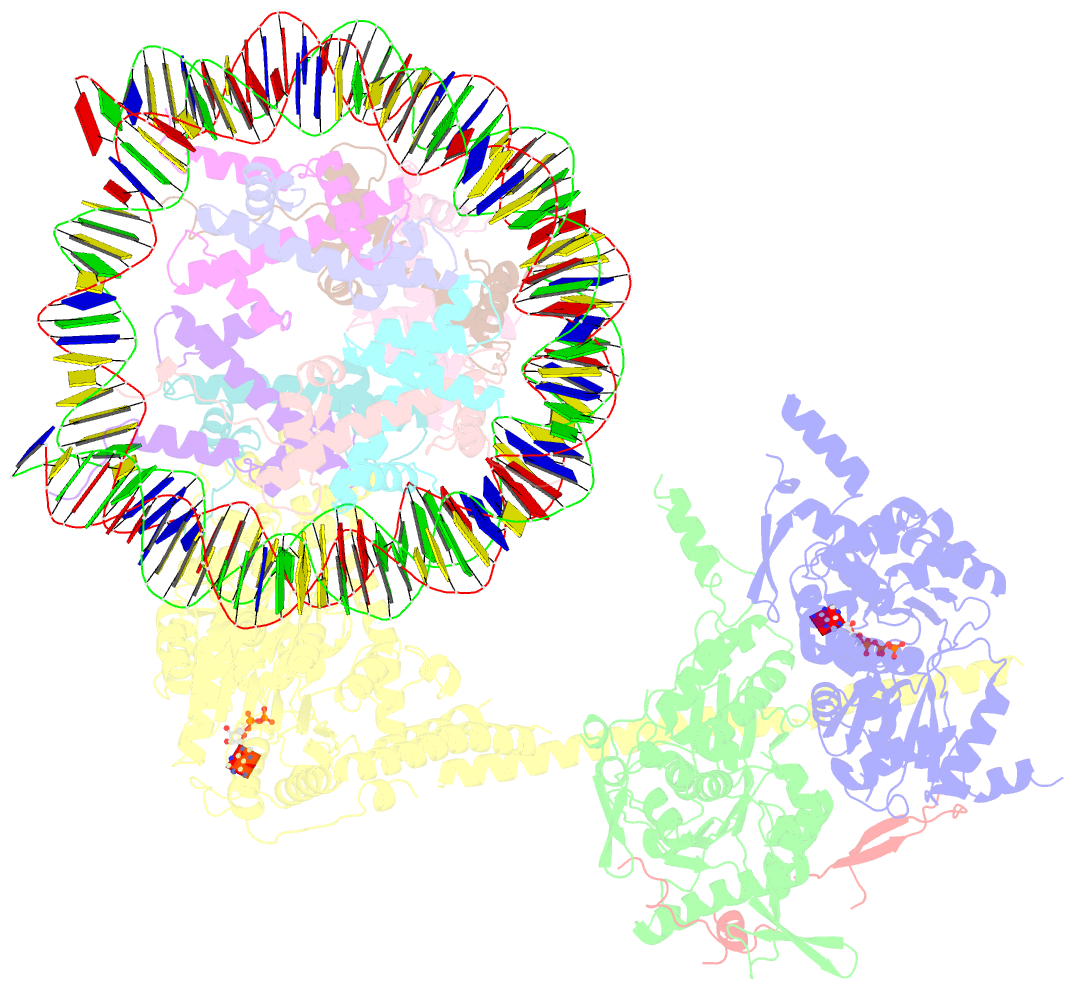
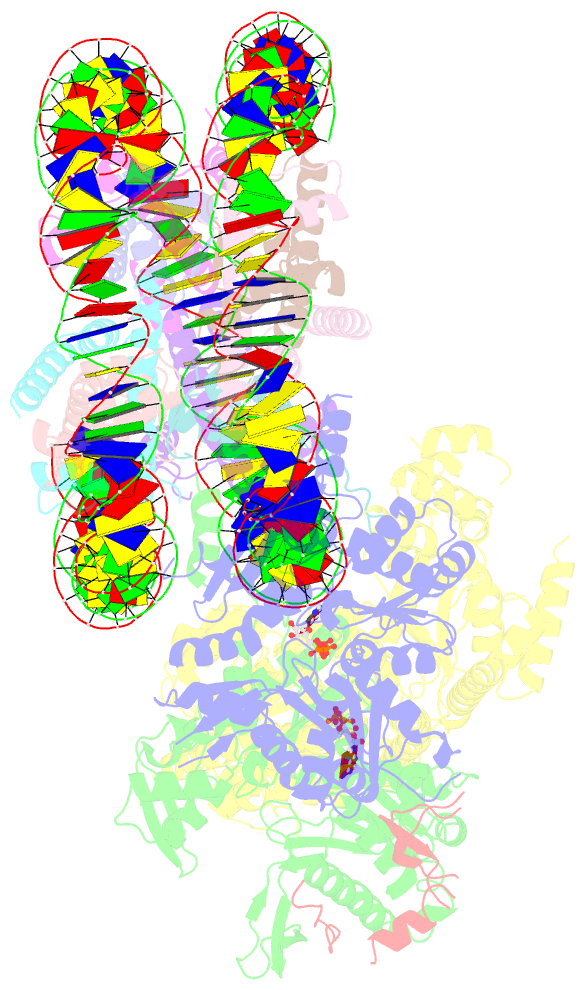
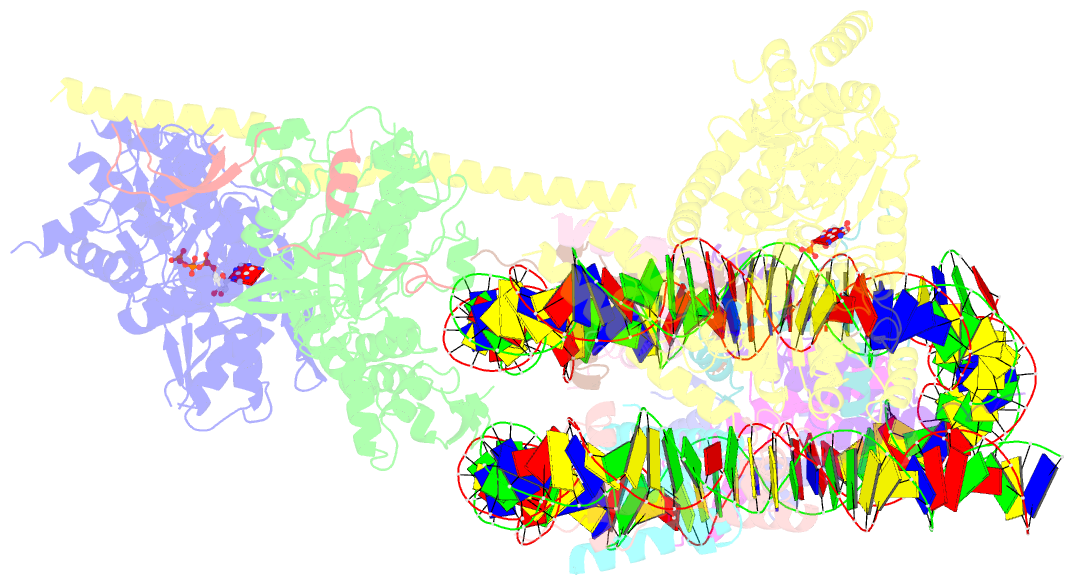
- cryo-EM structure of sth1-arp7-arp9-rtt102 bound to the nucleosome in adp beryllium fluoride state
- Reference
- Baker RW, Reimer JM, Carman PJ, Turegun B, Arakawa T, Dominguez R, Leschziner AE (2021): "Structural insights into assembly and function of the RSC chromatin remodeling complex." Nat.Struct.Mol.Biol., 28, 71-80. doi: 10.1038/s41594-020-00528-8.
- Abstract
- SWI/SNF chromatin remodelers modify the position and spacing of nucleosomes and, in humans, are linked to cancer. To provide insights into the assembly and regulation of this protein family, we focused on a subcomplex of the Saccharomyces cerevisiae RSC comprising its ATPase (Sth1), the essential actin-related proteins (ARPs) Arp7 and Arp9 and the ARP-binding protein Rtt102. Cryo-EM and biochemical analyses of this subcomplex shows that ARP binding induces a helical conformation in the helicase-SANT-associated (HSA) domain of Sth1. Surprisingly, the ARP module is rotated 120° relative to the full RSC about a pivot point previously identified as a regulatory hub in Sth1, suggesting that large conformational changes are part of Sth1 regulation and RSC assembly. We also show that a conserved interaction between Sth1 and the nucleosome acidic patch enhances remodeling. As some cancer-associated mutations dysregulate rather than inactivate SWI/SNF remodelers, our insights into RSC complex regulation advance a mechanistic understanding of chromatin remodeling in disease states.