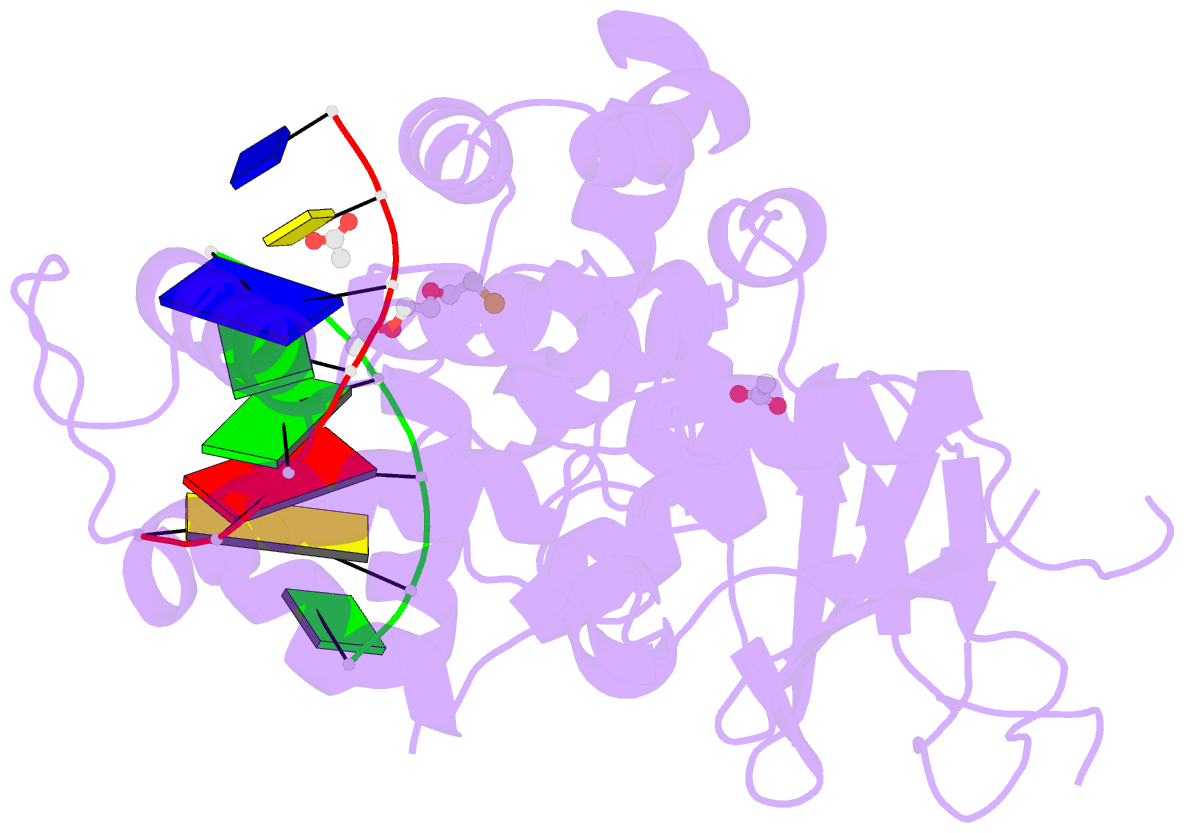
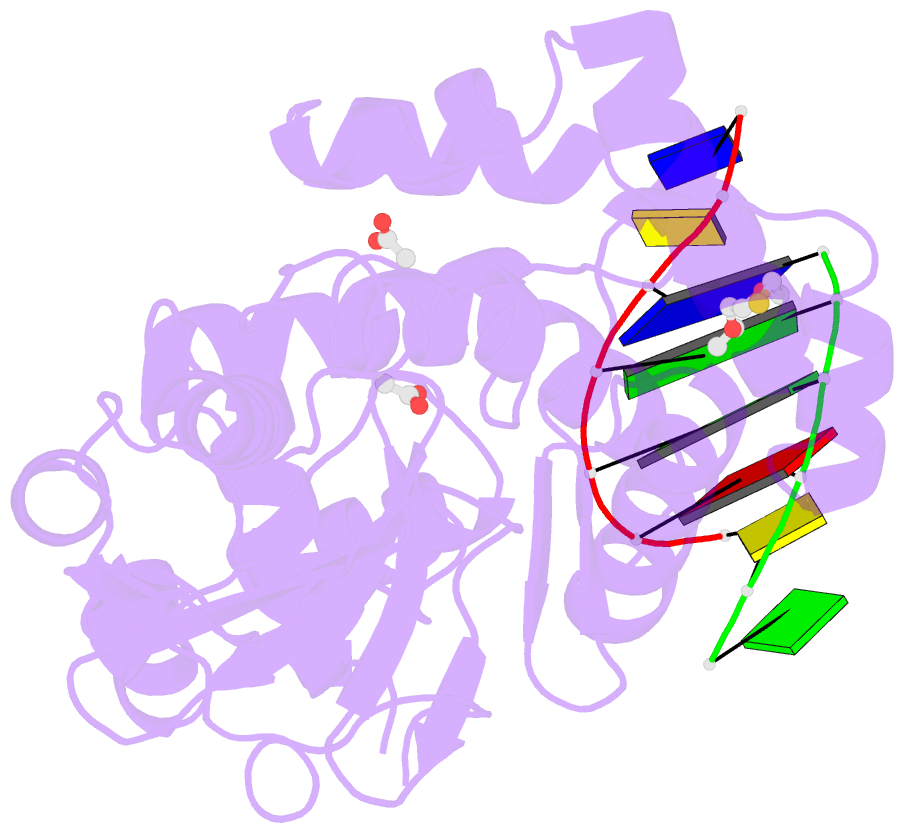
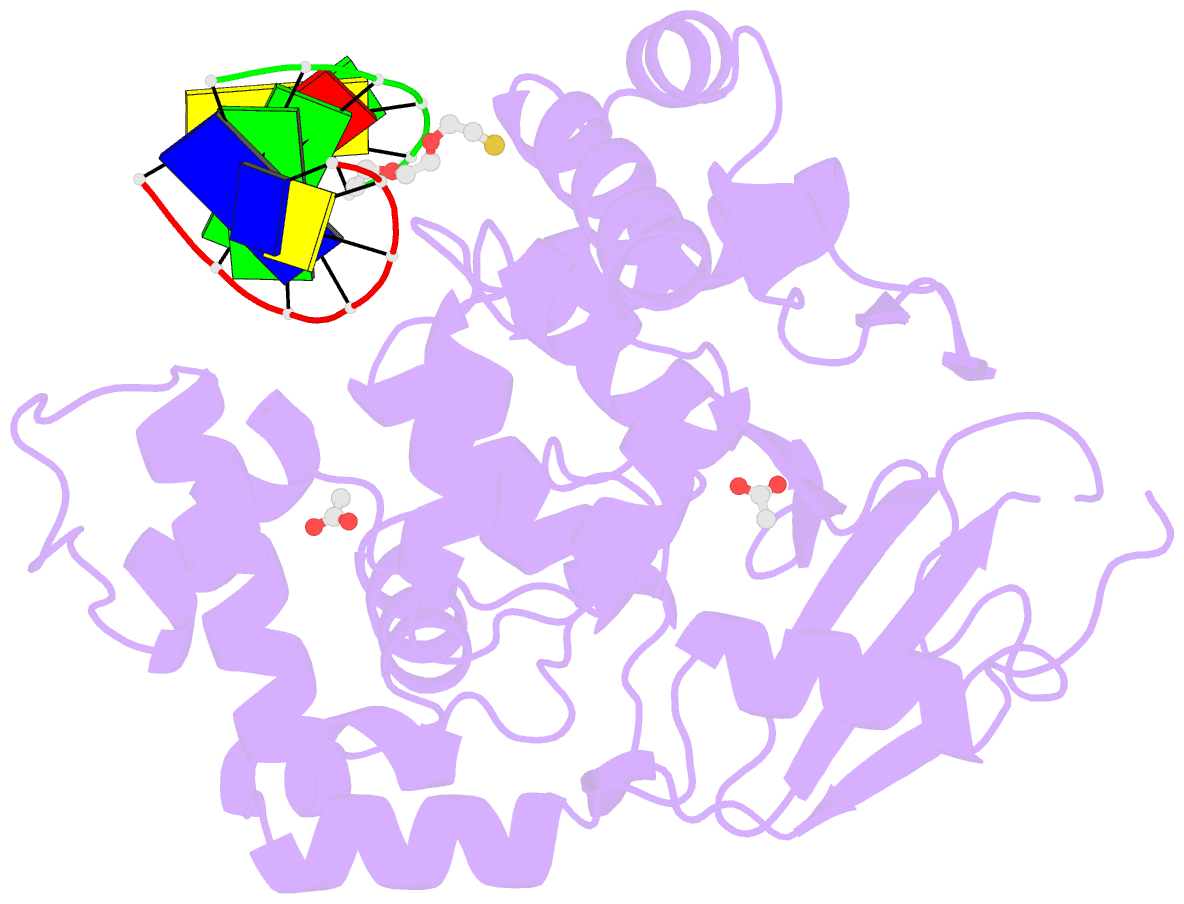
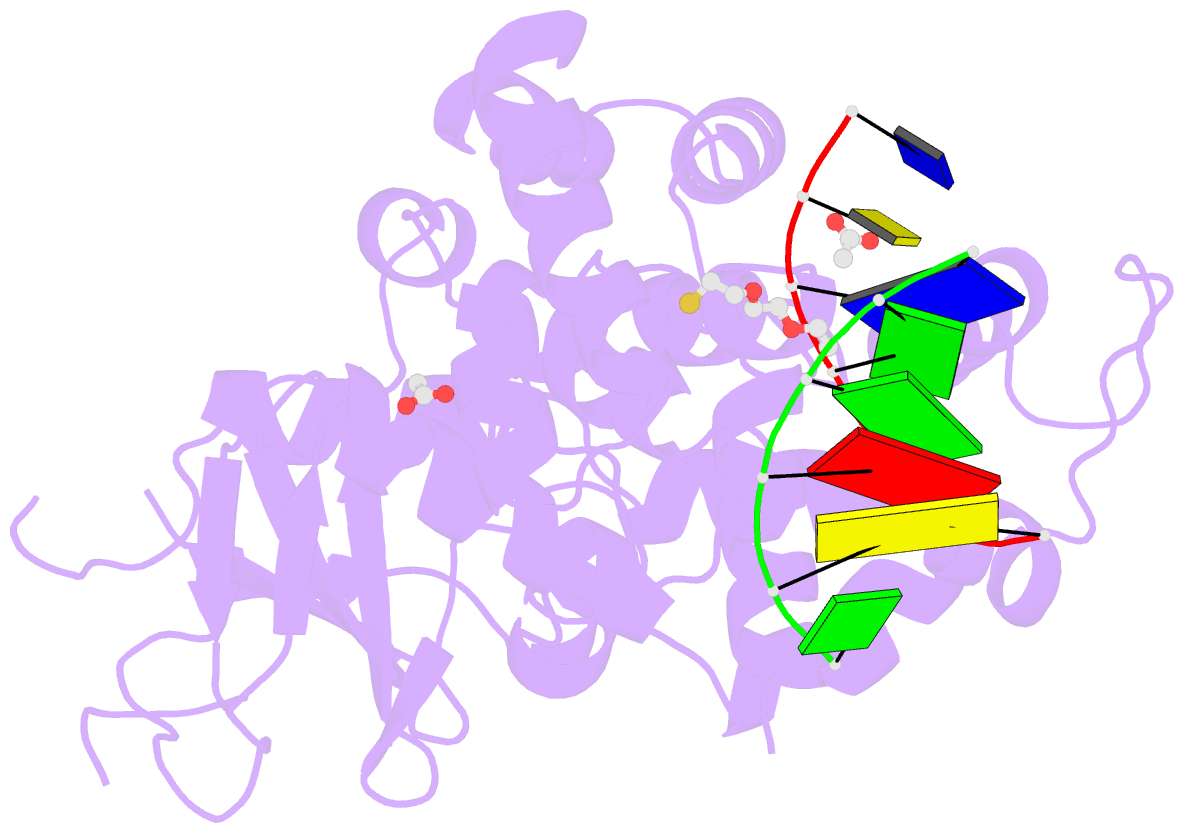
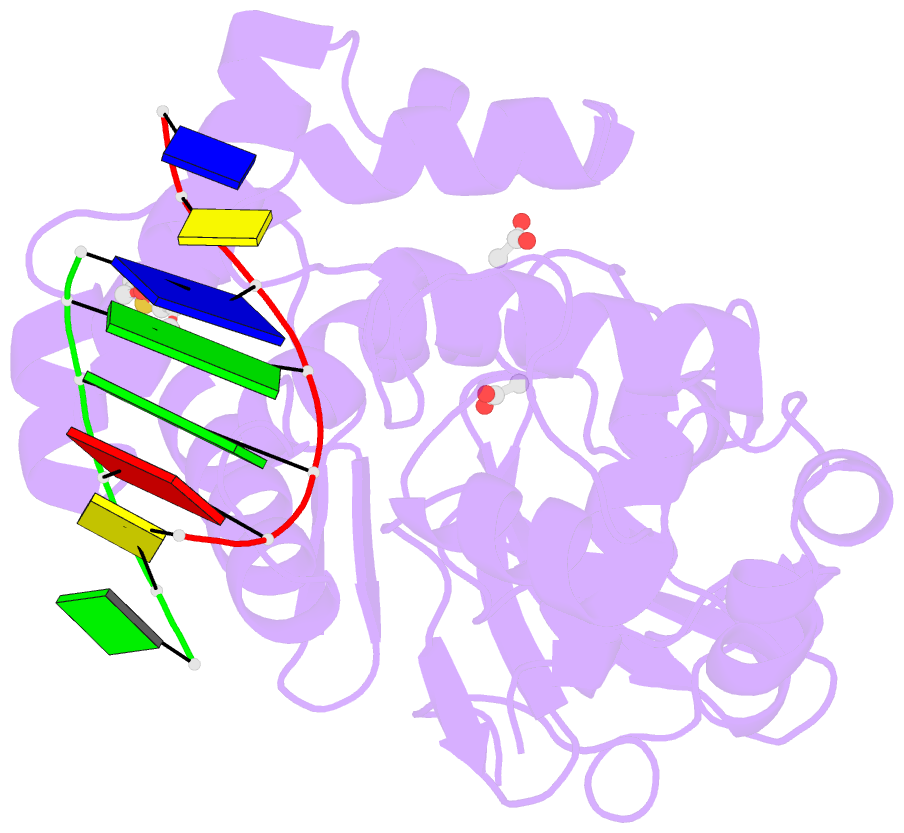
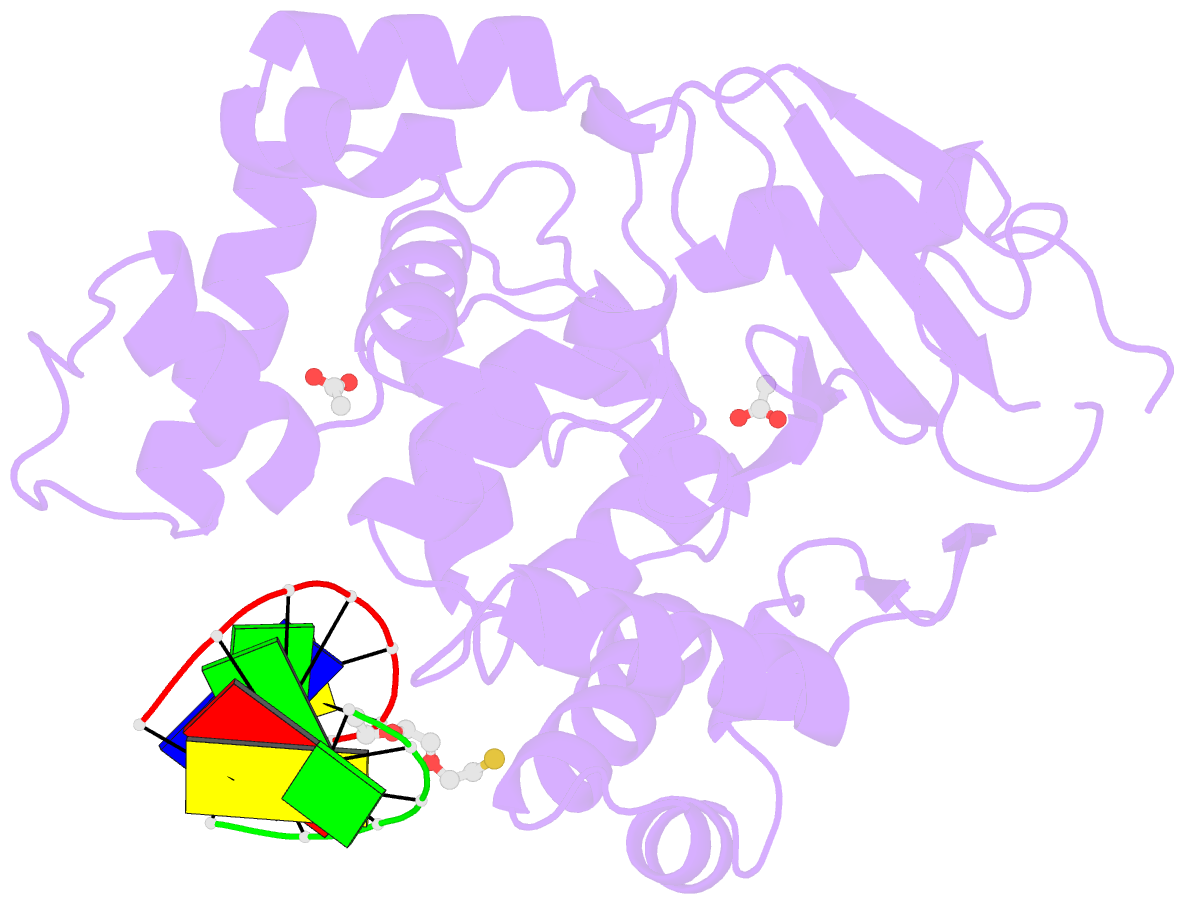
Summary information and primary citation
- PDB-id
- 6w13; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase-DNA
- Method
- X-ray (2.38 Å)
- Summary
- Human 8-oxoguanine glycosylase interrogating fully intrahelical oxog lesion DNA
- Reference
- Shigdel UK, Ovchinnikov V, Lee SJ, Shih JA, Karplus M, Nam K, Verdine GL (2020): "The trajectory of intrahelical lesion recognition and extrusion by the human 8-oxoguanine DNA glycosylase." Nat Commun, 11, 4437. doi: 10.1038/s41467-020-18290-2.
- Abstract
- Efficient search for DNA damage embedded in vast expanses of the DNA genome presents one of the greatest challenges to DNA repair enzymes. We report here crystal structures of human 8-oxoguanine (oxoG) DNA glycosylase, hOGG1, that interact with the DNA containing the damaged base oxoG and the normal base G while they are nested in the DNA helical stack. The structures reveal that hOGG1 engages the DNA using different protein-DNA contacts from those observed in the previously determined lesion recognition complex and other hOGG1-DNA complexes. By applying molecular dynamics simulations, we have determined the pathways taken by the lesion and normal bases when extruded from the DNA helix and their associated free energy profiles. These results reveal how the human oxoG DNA glycosylase hOGG1 locates the lesions inside the DNA helix and facilitates their extrusion for repair.