Summary information and primary citation
- PDB-id
- 6w6v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- cryo-EM (3.0 Å)
- Summary
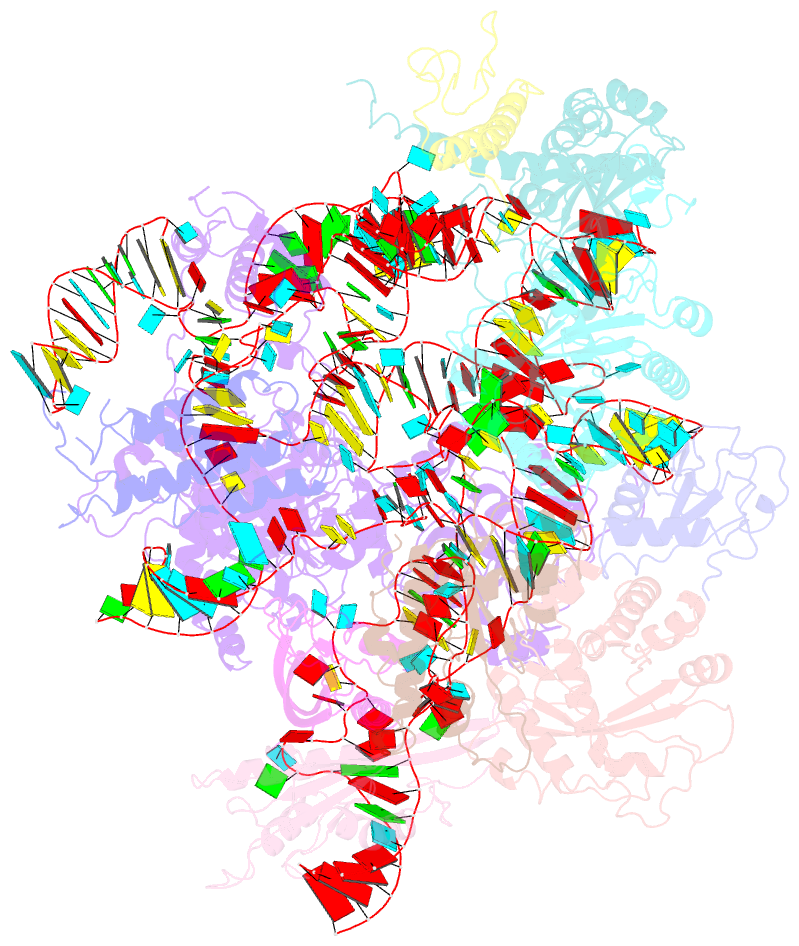
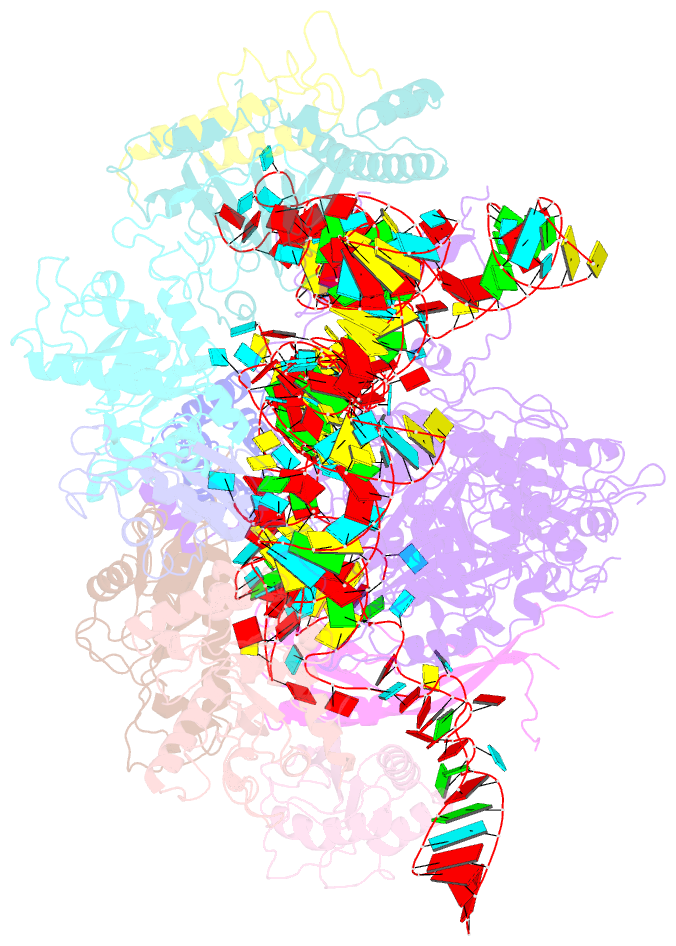
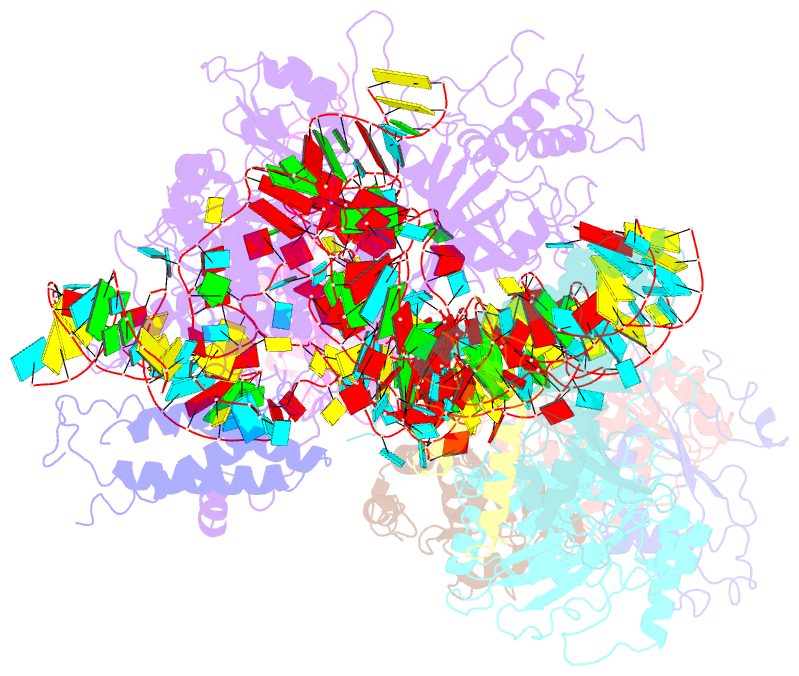
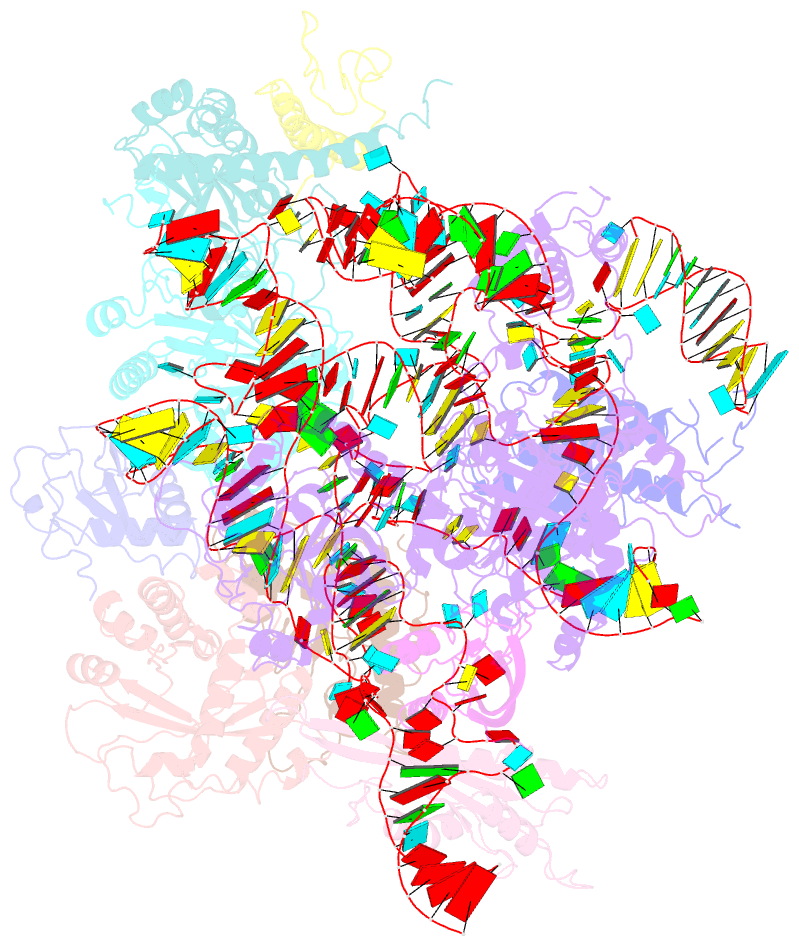
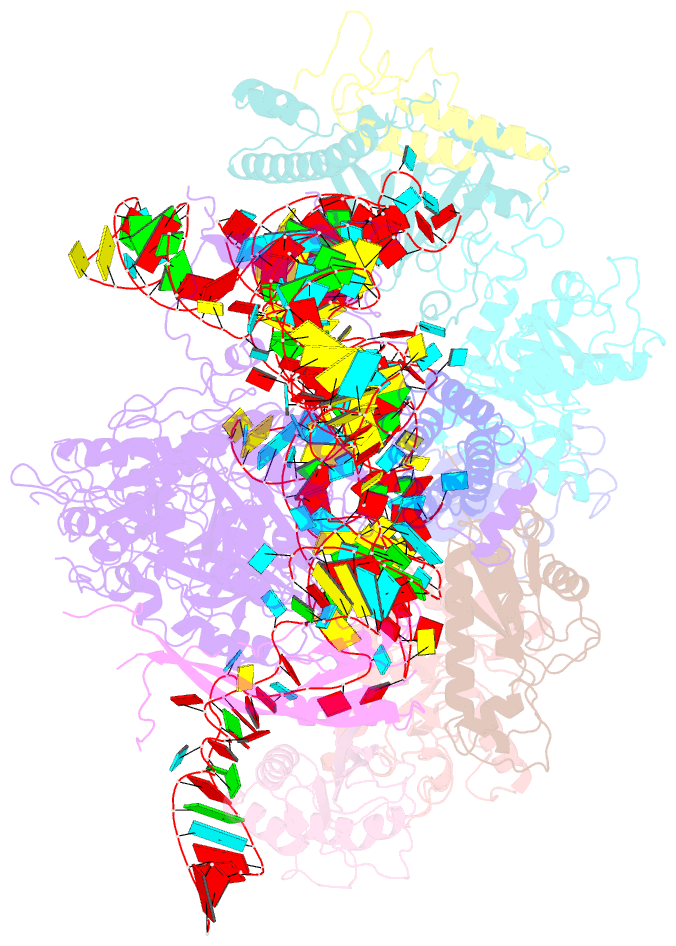
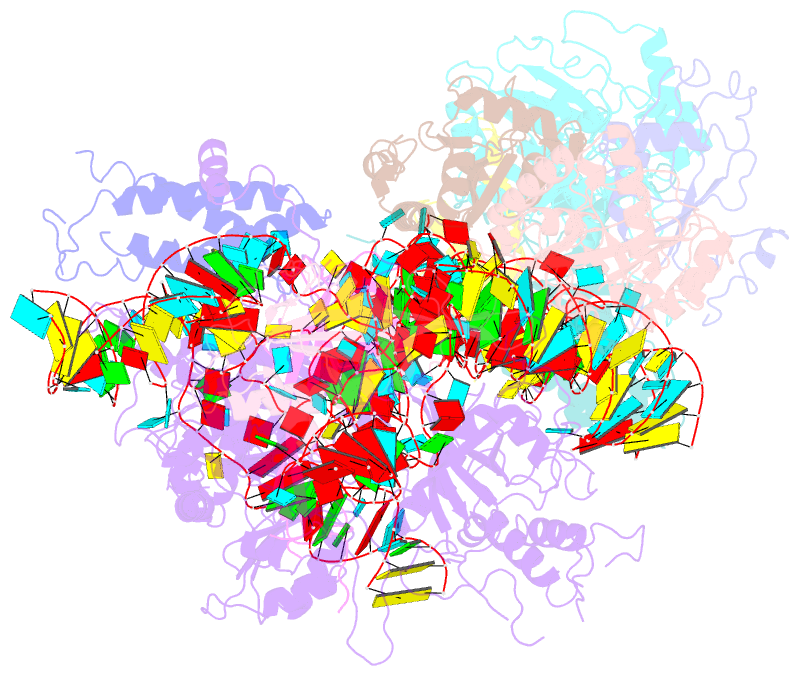
- Structure of yeast rnase mrp holoenzyme
- Reference
- Perederina A, Li D, Lee H, Bator C, Berezin I, Hafenstein SL, Krasilnikov AS (2020): "Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP." Nat Commun, 11, 3474. doi: 10.1038/s41467-020-17308-z.
- Abstract
- RNase MRP is an essential eukaryotic ribonucleoprotein complex involved in the maturation of rRNA and the regulation of the cell cycle. RNase MRP is related to the ribozyme-based RNase P, but it has evolved to have distinct cellular roles. We report a cryo-EM structure of the S. cerevisiae RNase MRP holoenzyme solved to 3.0 Å. We describe the structure of this 450 kDa complex, interactions between its components, and the organization of its catalytic RNA. We show that some of the RNase MRP proteins shared with RNase P undergo an unexpected RNA-driven remodeling that allows them to bind to divergent RNAs. Further, we reveal how this RNA-driven protein remodeling, acting together with the introduction of new auxiliary elements, results in the functional diversification of RNase MRP and its progenitor, RNase P, and demonstrate structural underpinnings of the acquisition of new functions by catalytic RNPs.