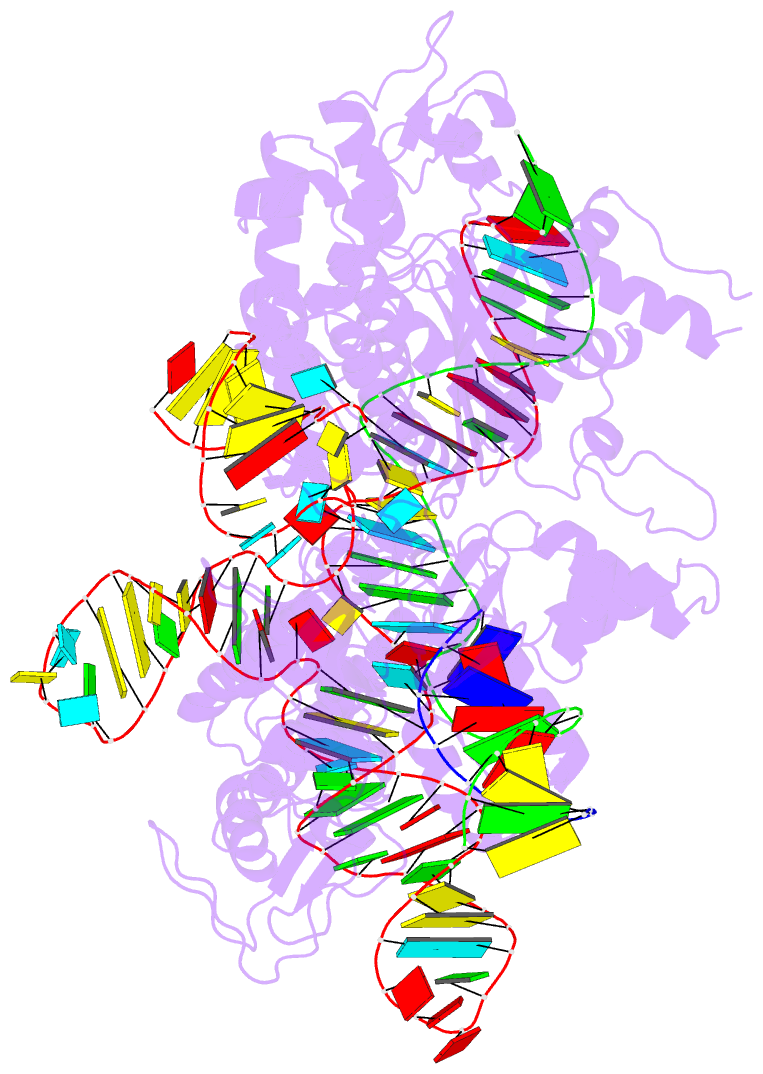
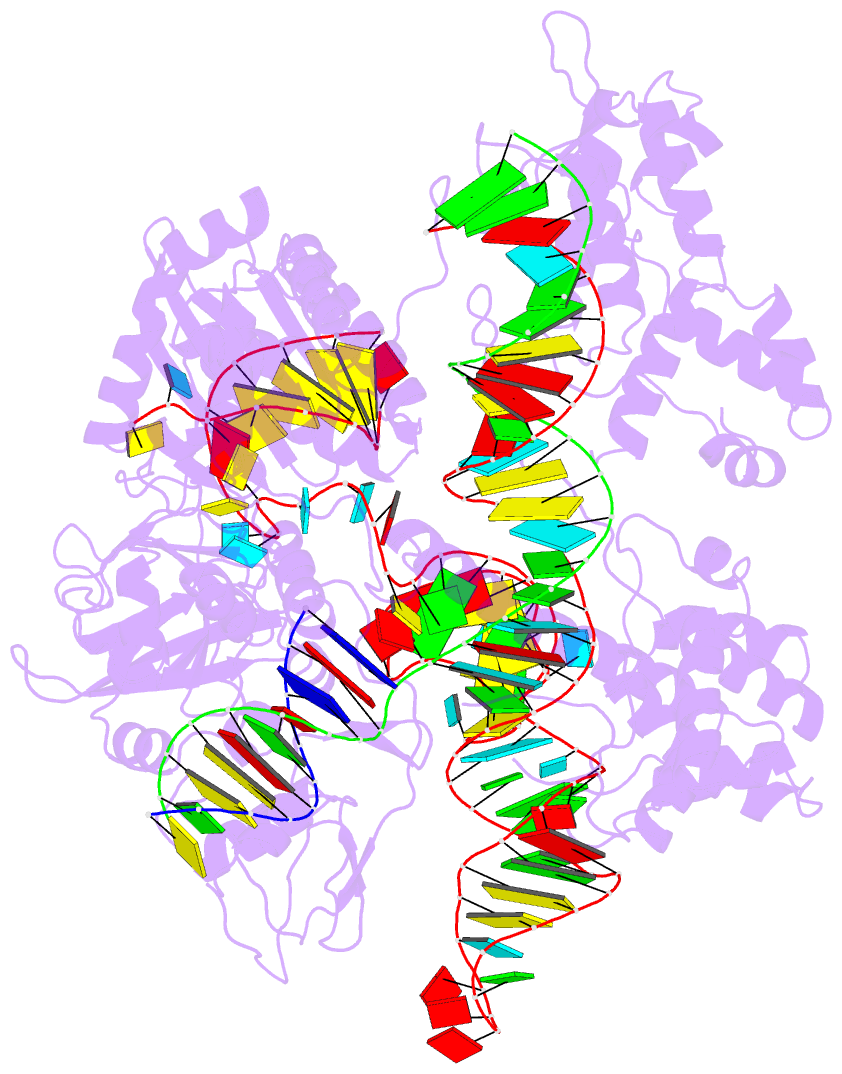
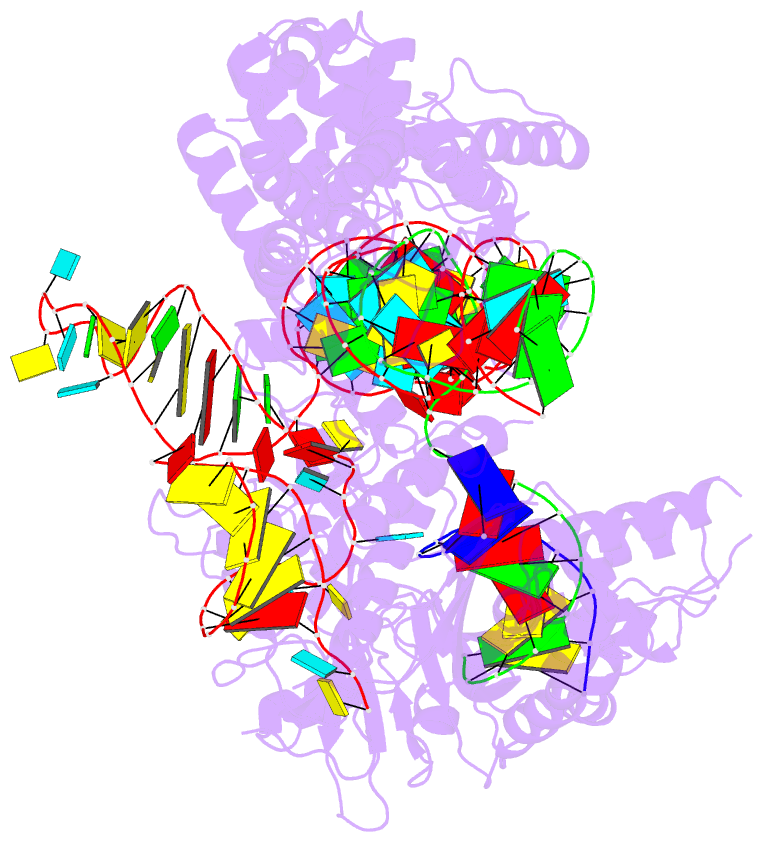
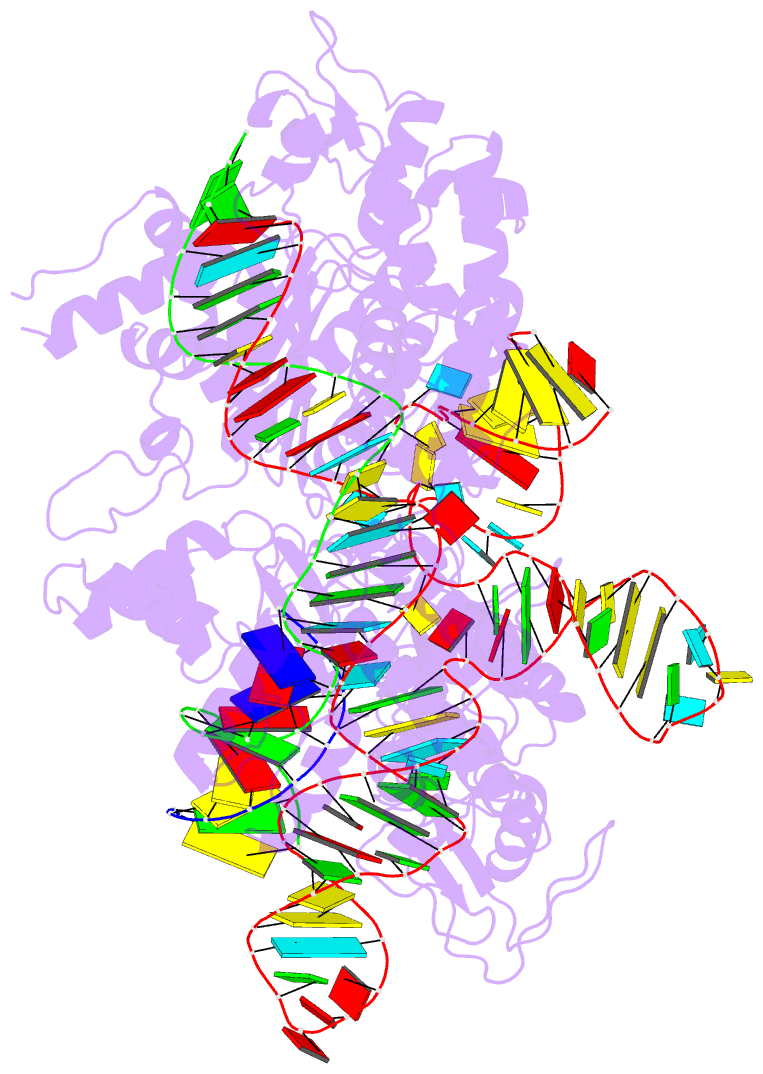
Summary information and primary citation
- PDB-id
- 6wc0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (3.61 Å)
- Summary
- Crystal structure of acecas9 bound with guide RNA and DNA with 5'-nnntc-3' pam
- Reference
- Das A, Hand TH, Smith CL, Wickline E, Zawrotny M, Li H (2020): "The molecular basis for recognition of 5'-NNNCC-3' PAM and its methylation state by Acidothermus cellulolyticus Cas9." Nat Commun, 11, 6346. doi: 10.1038/s41467-020-20204-1.
- Abstract
- Acidothermus cellulolyticus CRISPR-Cas9 (AceCas9) is a thermophilic Type II-C enzyme that has potential genome editing applications in extreme environments. It cleaves DNA with a 5'-NNNCC-3' Protospacer Adjacent Motif (PAM) and is sensitive to its methylation status. To understand the molecular basis for the high specificity of AceCas9 for its PAM, we determined two crystal structures of AceCas9 lacking its HNH domain (AceCas9-ΔHNH) bound with a single guide RNA and DNA substrates, one with the correct and the other with an incorrect PAM. Three residues, Glu1044, Arg1088, Arg1091, form an intricate hydrogen bond network with the first cytosine and the two opposing guanine nucleotides to confer specificity. Methylation of the first but not the second cytosine base abolishes AceCas9 activity, consistent with the observed PAM recognition pattern. The high sensitivity of AceCas9 to the modified cytosine makes it a potential device for detecting epigenomic changes in genomes.