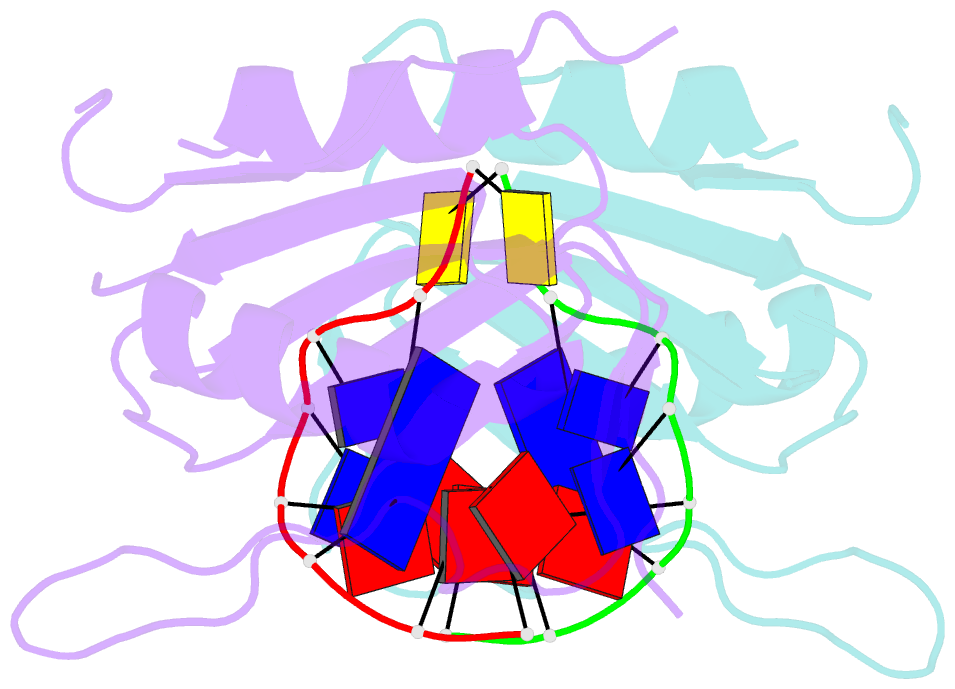
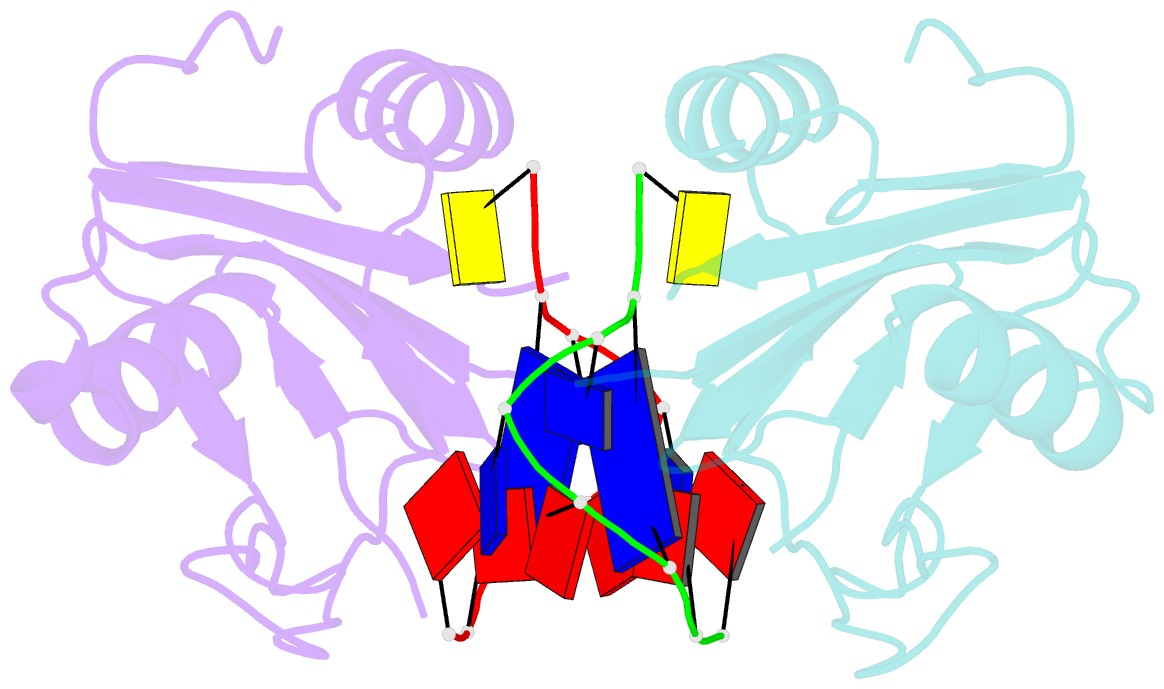
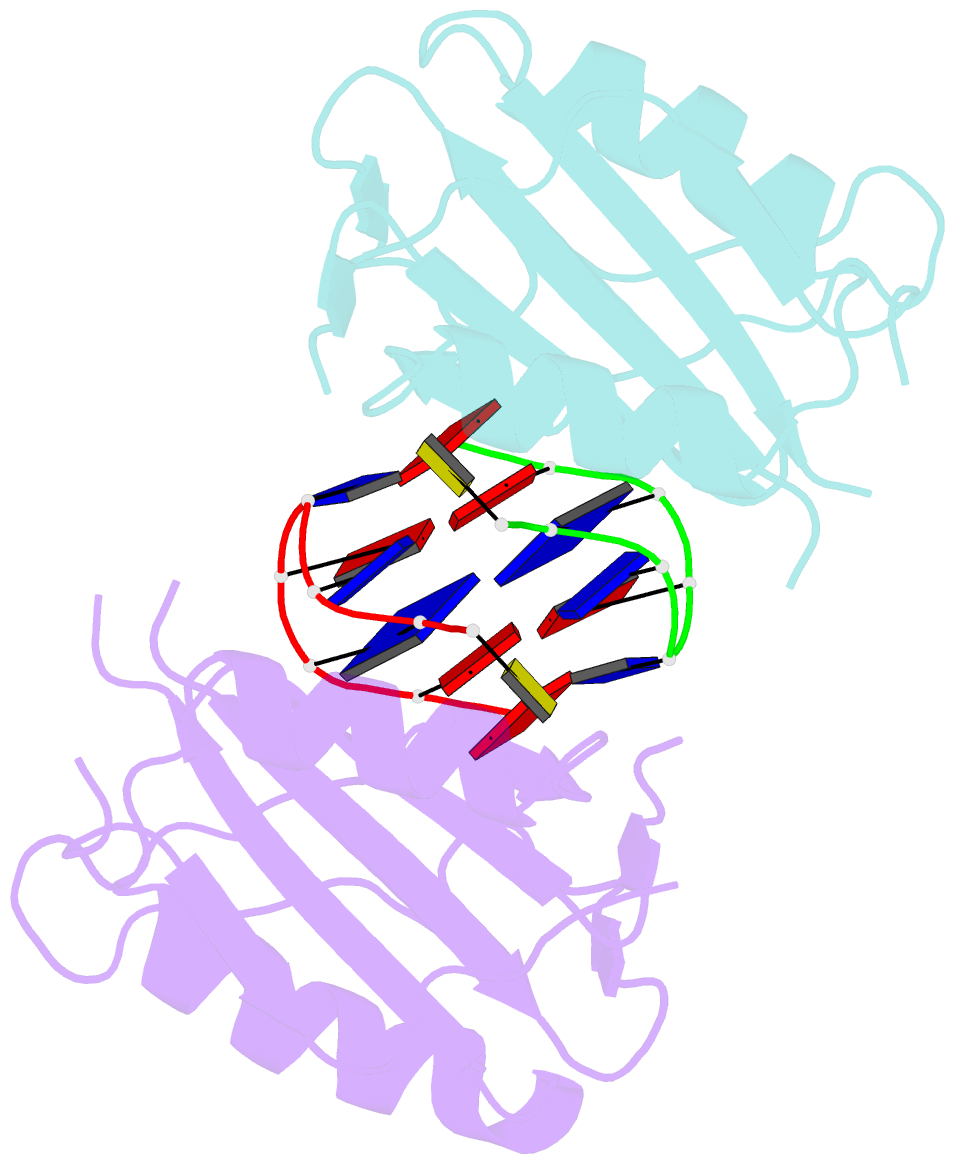
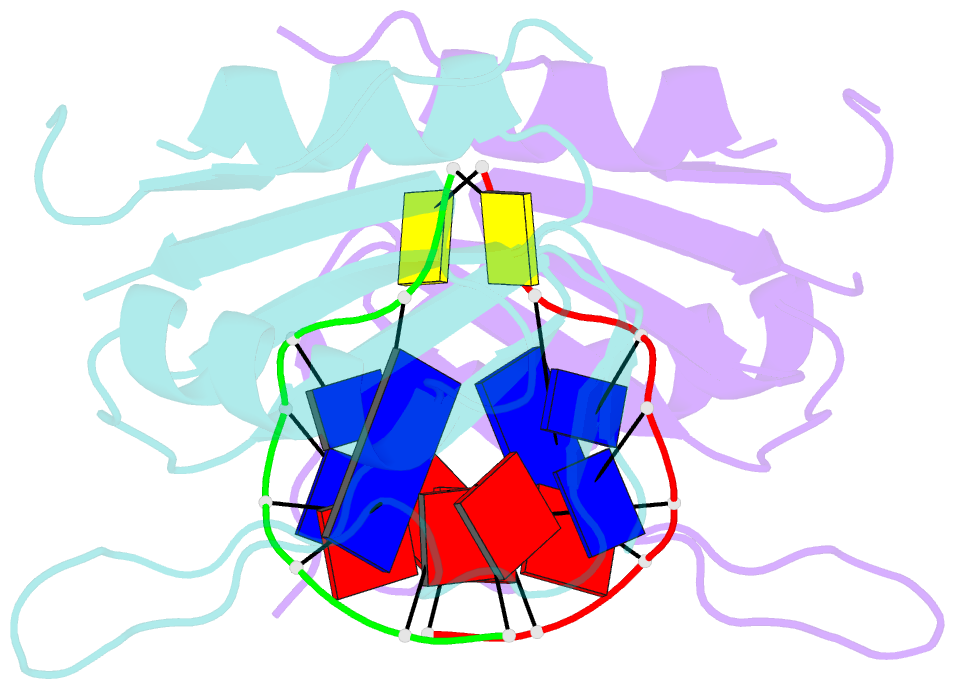
Summary information and primary citation
- PDB-id
- 6we1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.612 Å)
- Summary
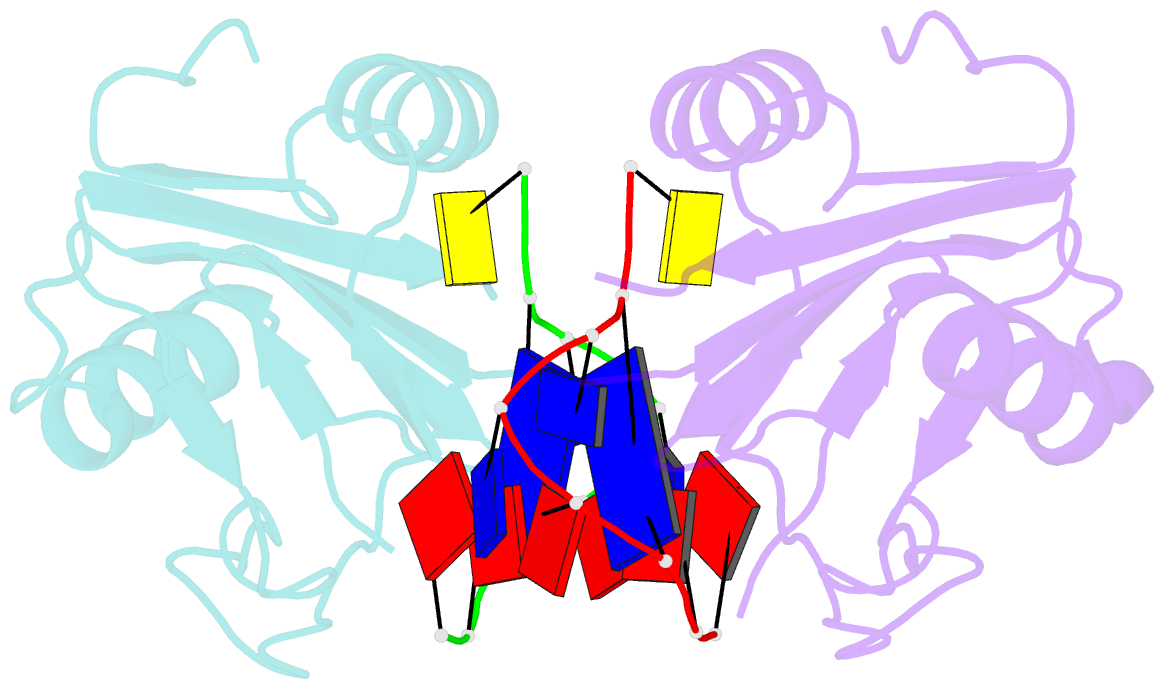
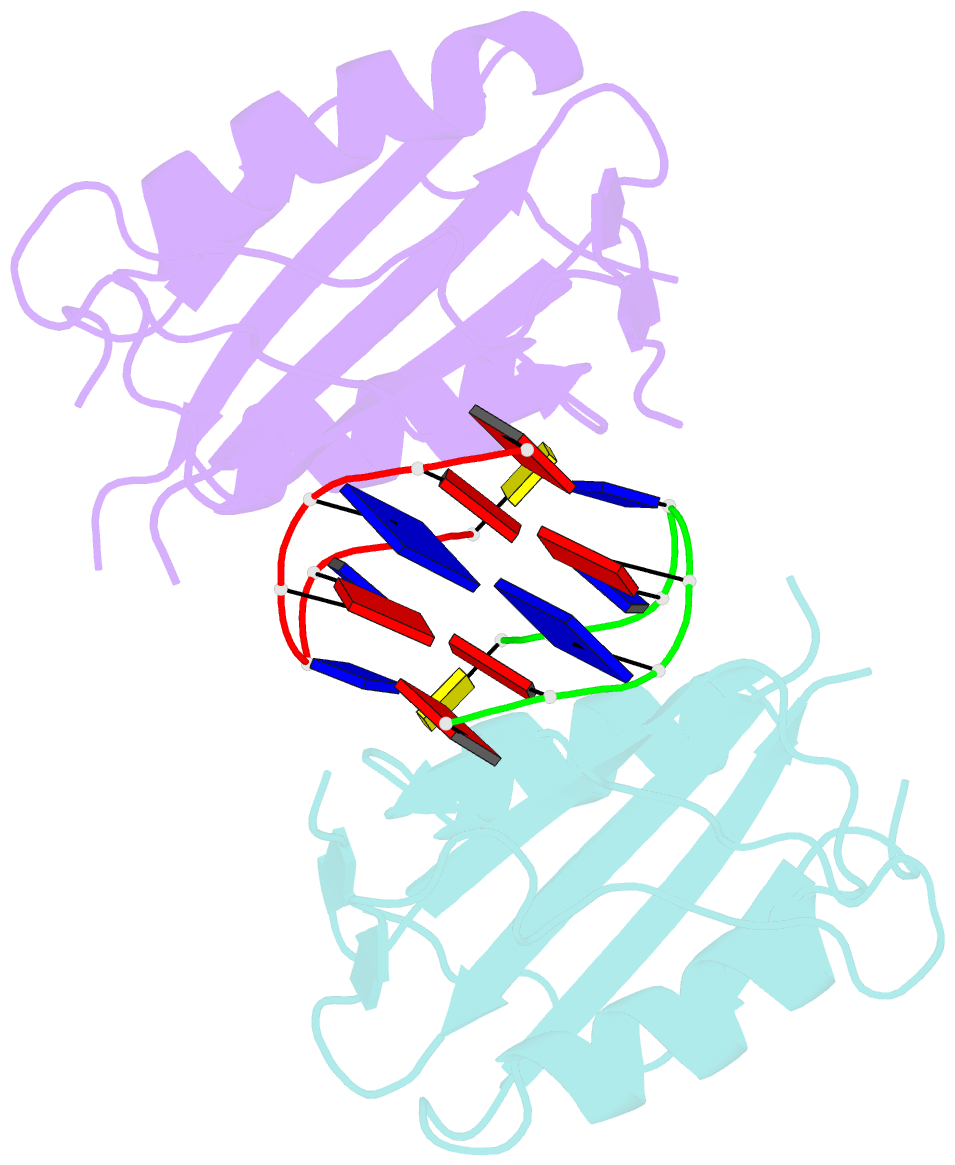
- Wheat dwarf virus rep domain complexed with a single-stranded DNA 8-mer comprising the cleavage site
- Reference
- Tompkins KJ, Houtti M, Litzau LA, Aird EJ, Everett BA, Nelson AT, Pornschloegl L, Limon-Swanson LK, Evans RL, Evans K, Shi K, Aihara H, Gordon WR (2021): "Molecular underpinnings of ssDNA specificity by Rep HUH-endonucleases and implications for HUH-tag multiplexing and engineering." Nucleic Acids Res., 49, 1046-1064. doi: 10.1093/nar/gkaa1248.
- Abstract
- Replication initiator proteins (Reps) from the HUH-endonuclease superfamily process specific single-stranded DNA (ssDNA) sequences to initiate rolling circle/hairpin replication in viruses, such as crop ravaging geminiviruses and human disease causing parvoviruses. In biotechnology contexts, Reps are the basis for HUH-tag bioconjugation and a critical adeno-associated virus genome integration tool. We solved the first co-crystal structures of Reps complexed to ssDNA, revealing a key motif for conferring sequence specificity and for anchoring a bent DNA architecture. In combination, we developed a deep sequencing cleavage assay, termed HUH-seq, to interrogate subtleties in Rep specificity and demonstrate how differences can be exploited for multiplexed HUH-tagging. Together, our insights allowed engineering of only four amino acids in a Rep chimera to predictably alter sequence specificity. These results have important implications for modulating viral infections, developing Rep-based genomic integration tools, and enabling massively parallel HUH-tag barcoding and bioconjugation applications.