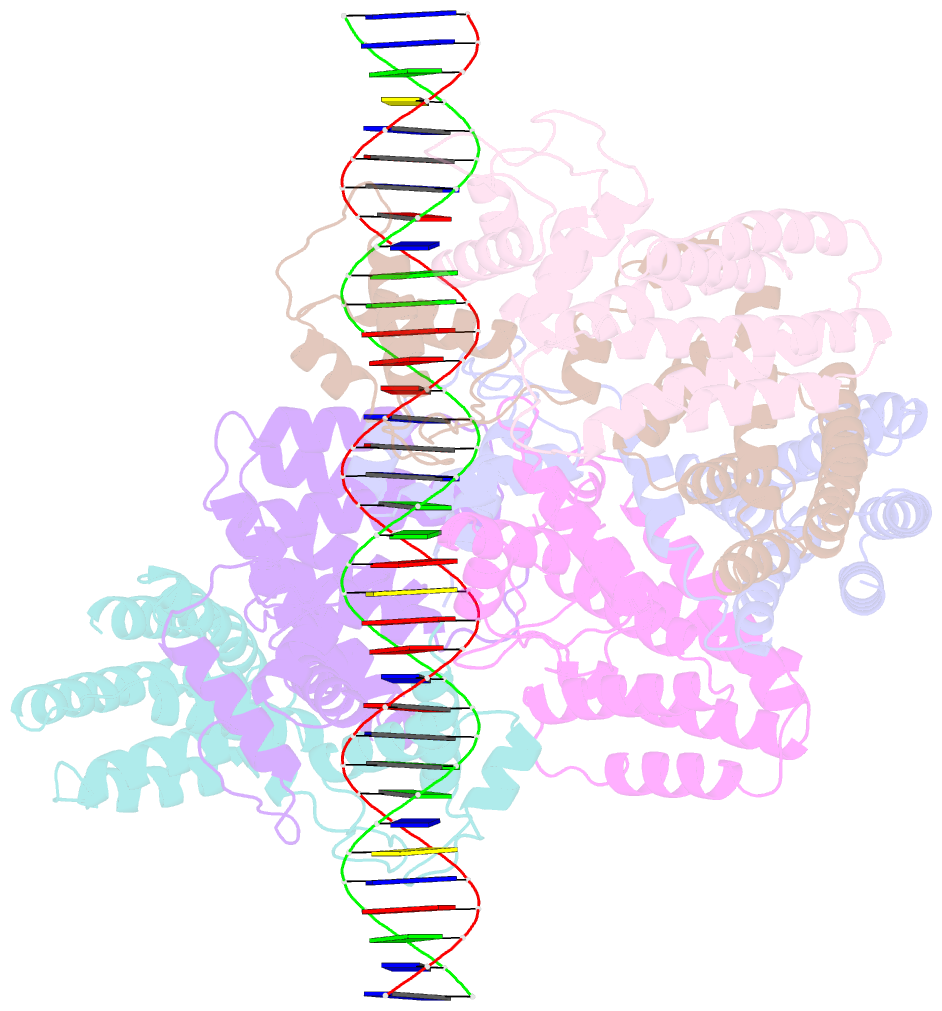
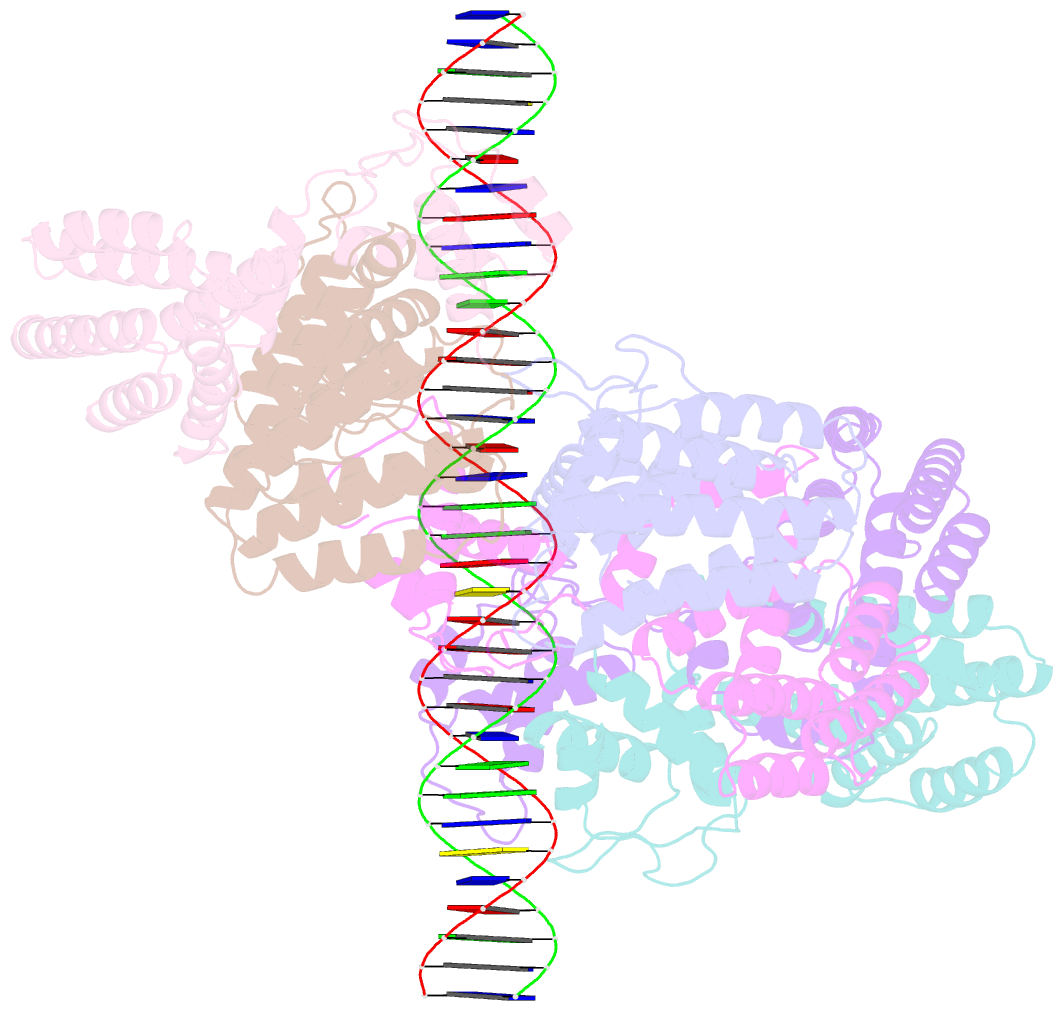
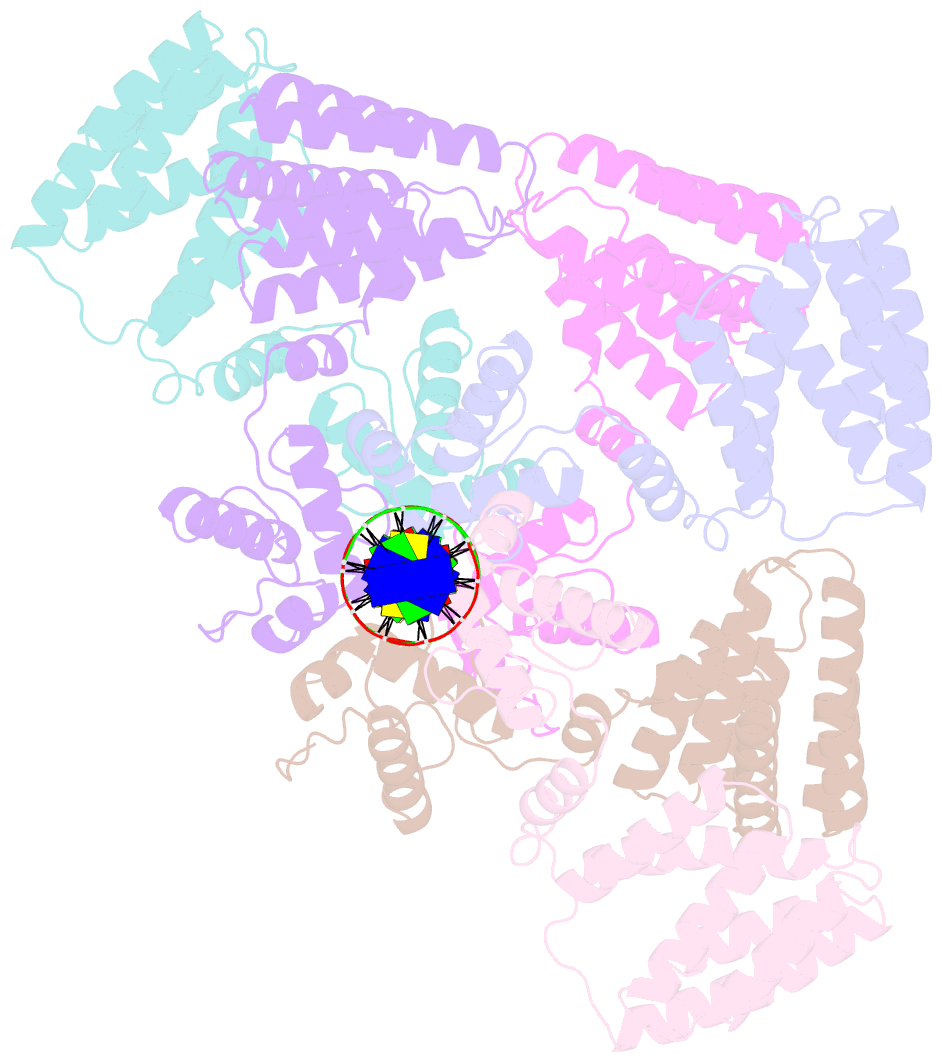
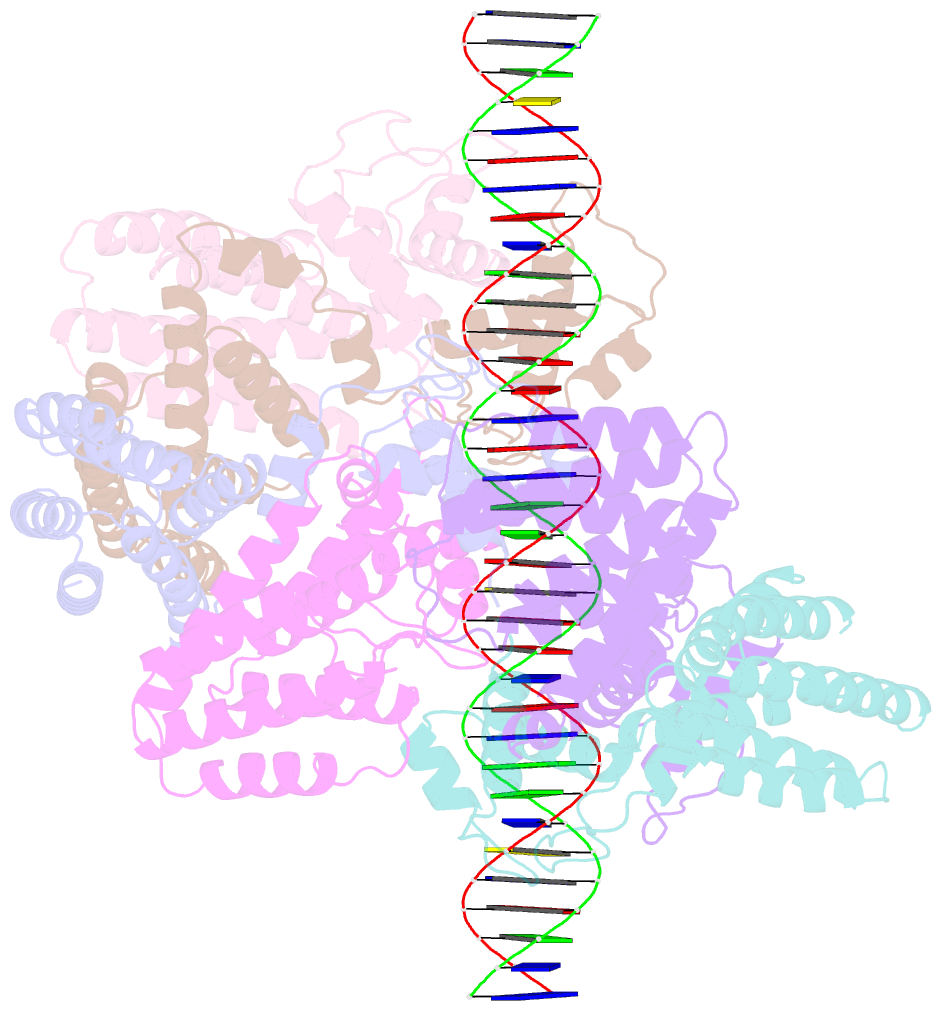
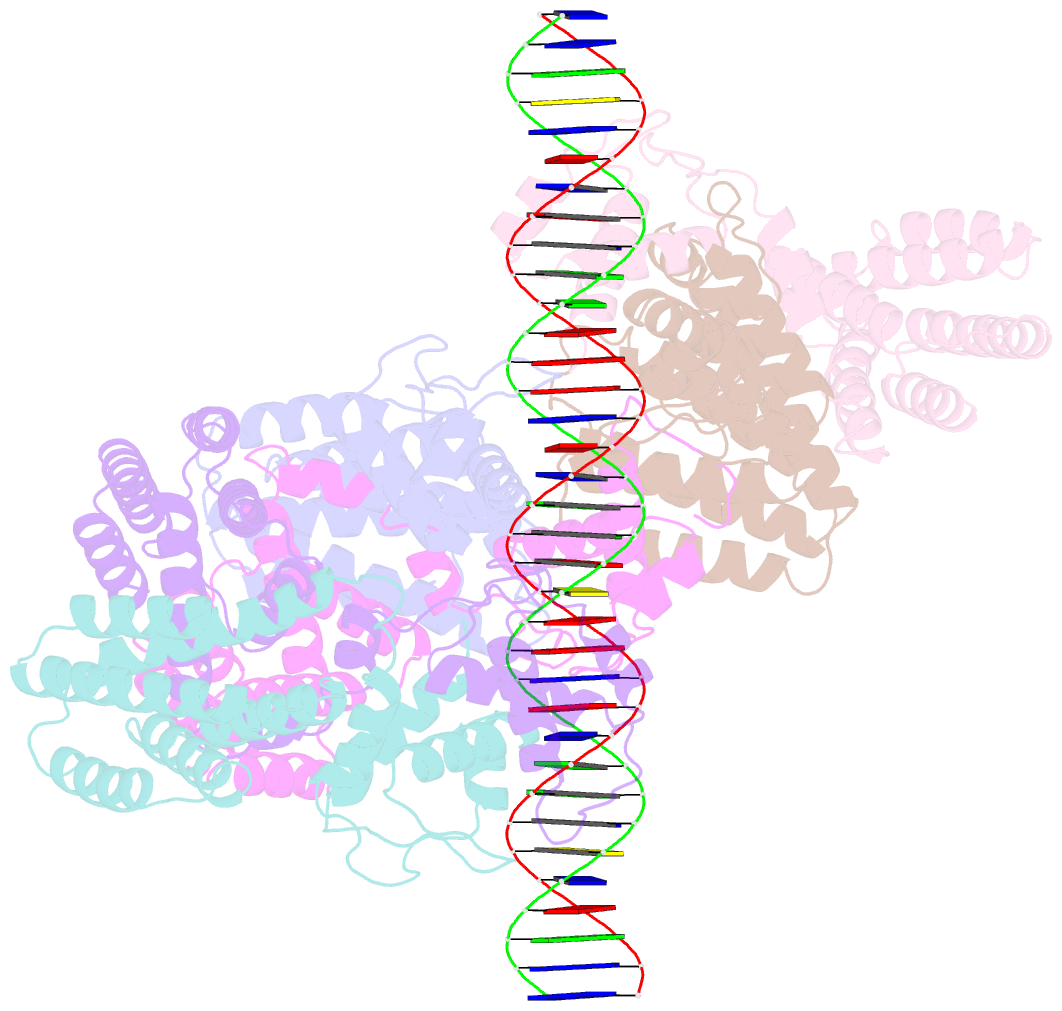
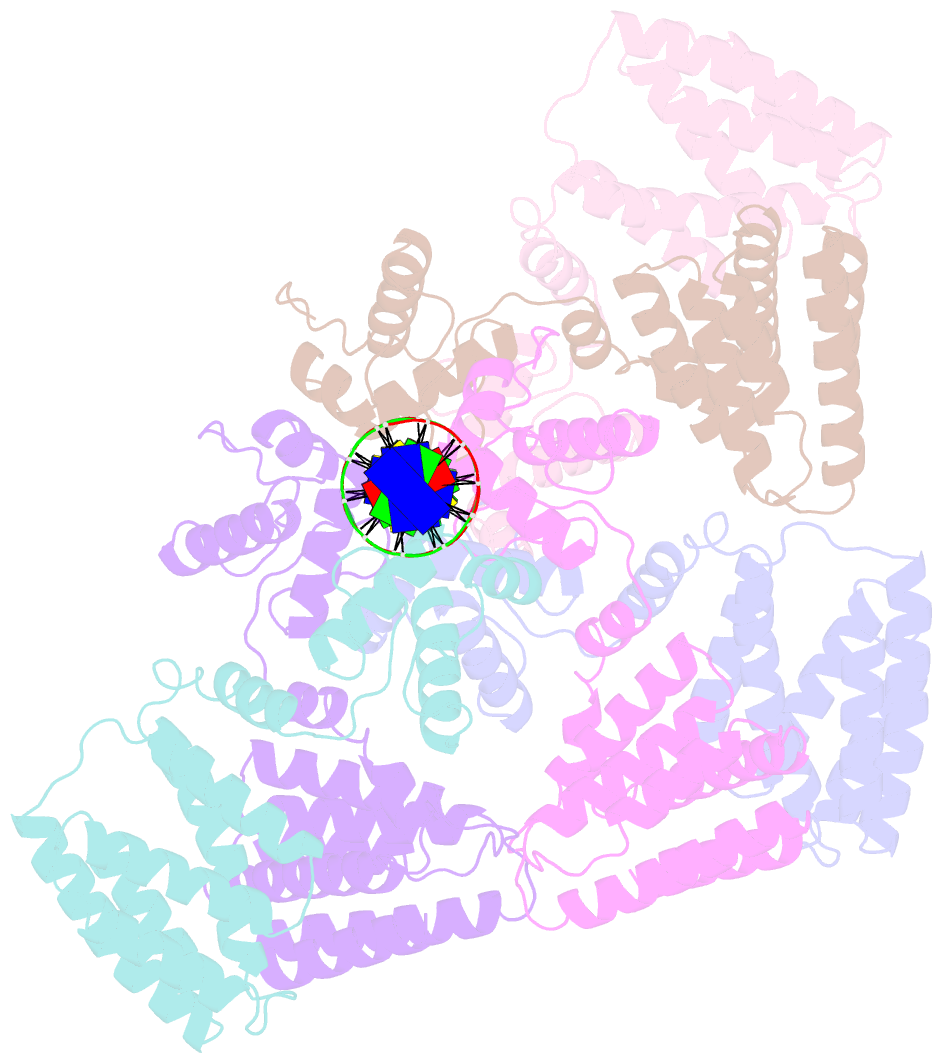
Summary information and primary citation
- PDB-id
- 6wg7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (8.3 Å)
- Summary
- Coordinates of nanr dimer fitted in hexameric nanr-DNA hetero-complex cryo-EM map
- Reference
- Horne CR, Venugopal H, Panjikar S, Wood DM, Henrickson A, Brookes E, North RA, Murphy JM, Friemann R, Griffin MDW, Ramm G, Demeler B, Dobson RCJ (2021): "Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism." Nat Commun, 12, 1988. doi: 10.1038/s41467-021-22253-6.
- Abstract
- Bacteria respond to environmental changes by inducing transcription of some genes and repressing others. Sialic acids, which coat human cell surfaces, are a nutrient source for pathogenic and commensal bacteria. The Escherichia coli GntR-type transcriptional repressor, NanR, regulates sialic acid metabolism, but the mechanism is unclear. Here, we demonstrate that three NanR dimers bind a (GGTATA)3-repeat operator cooperatively and with high affinity. Single-particle cryo-electron microscopy structures reveal the DNA-binding domain is reorganized to engage DNA, while three dimers assemble in close proximity across the (GGTATA)3-repeat operator. Such an interaction allows cooperative protein-protein interactions between NanR dimers via their N-terminal extensions. The effector, N-acetylneuraminate, binds NanR and attenuates the NanR-DNA interaction. The crystal structure of NanR in complex with N-acetylneuraminate reveals a domain rearrangement upon N-acetylneuraminate binding to lock NanR in a conformation that weakens DNA binding. Our data provide a molecular basis for the regulation of bacterial sialic acid metabolism.