Summary information and primary citation
- PDB-id
- 6wpi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.02 Å)
- Summary
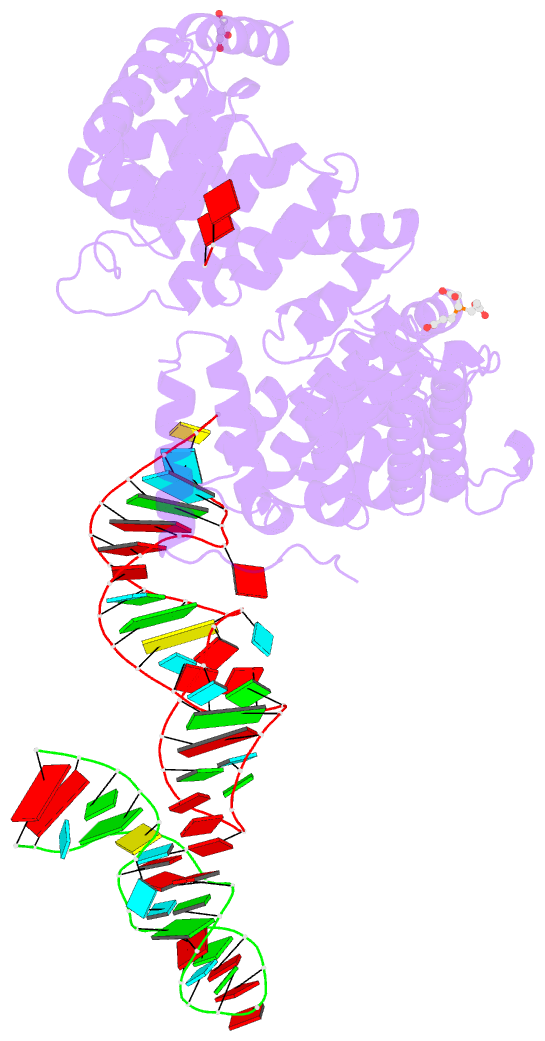
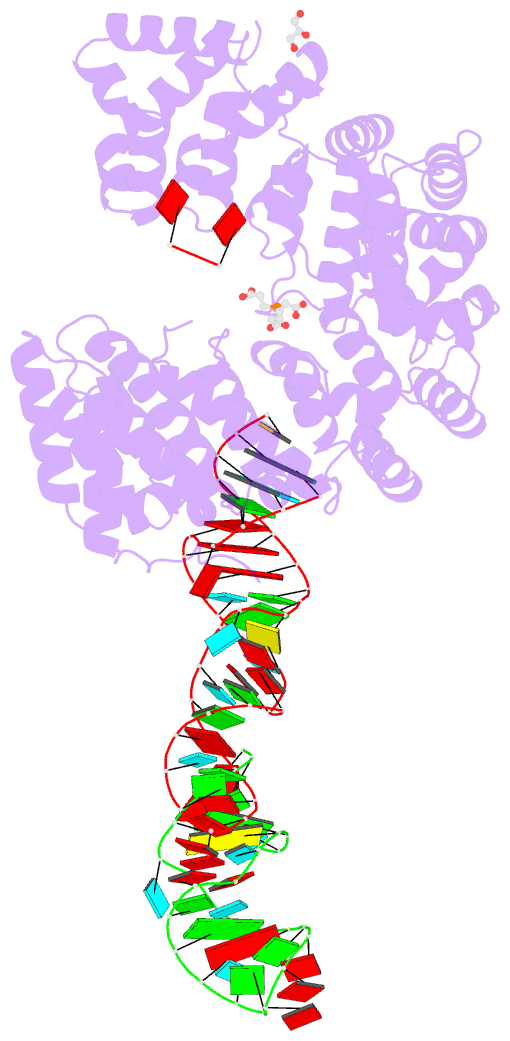
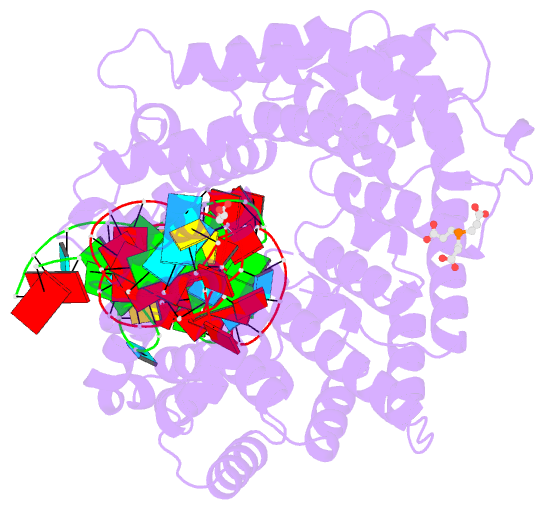
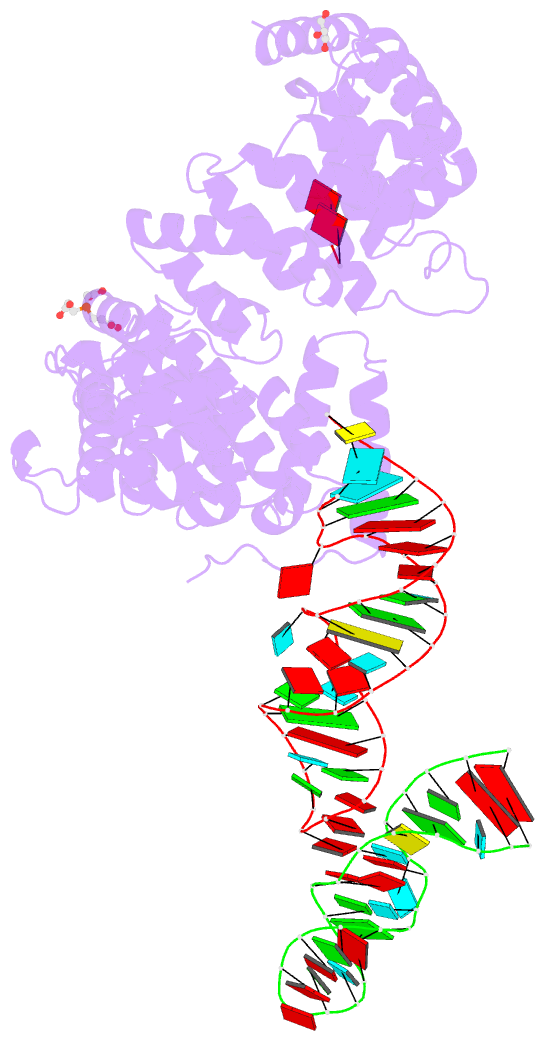
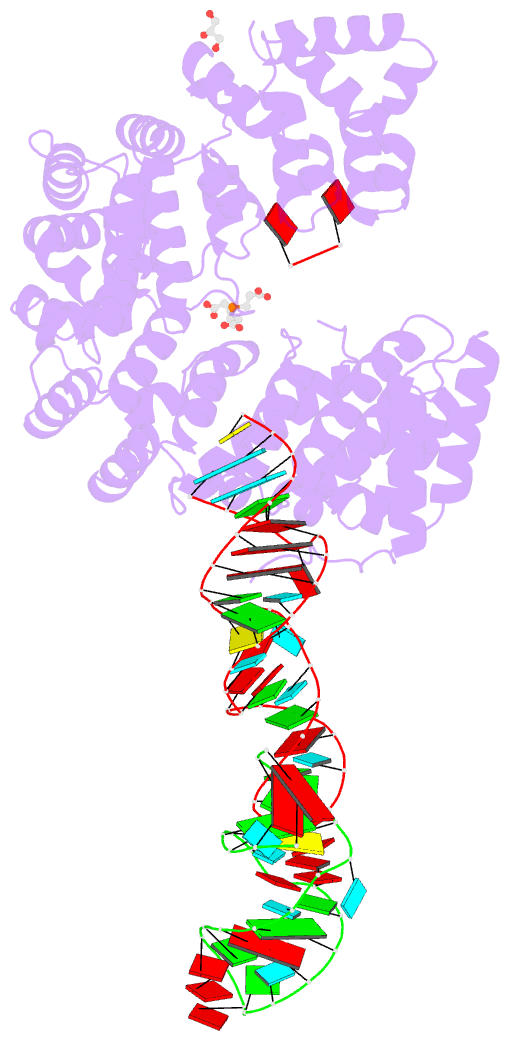
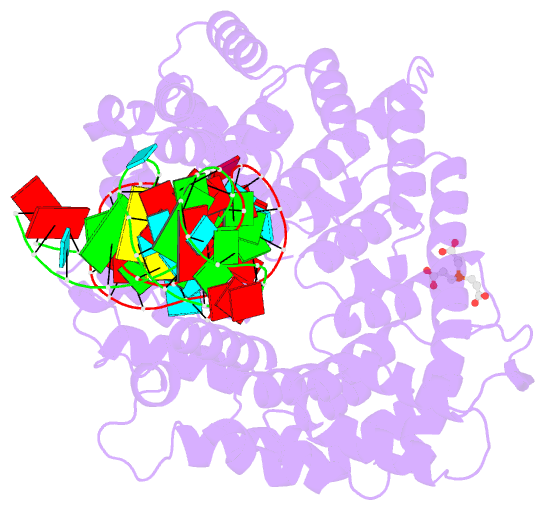
- Crystal structure of nop9 in complex with its1 RNA
- Reference
- Zhang J, Teramoto T, Qiu C, Wine RN, Gonzalez LE, Baserga SJ, Tanaka Hall TM (2020): "Nop9 recognizes structured and single-stranded RNA elements of preribosomal RNA." Rna, 26, 1049-1059. doi: 10.1261/rna.075416.120.
- Abstract
- Nop9 is an essential factor in the processing of preribosomal RNA. Its absence in yeast is lethal, and defects in the human ortholog are associated with breast cancer, autoimmunity, and learning/language impairment. PUF family RNA-binding proteins are best known for sequence-specific RNA recognition, and most contain eight α-helical repeats that bind to the RNA bases of single-stranded RNA. Nop9 is an unusual member of this family in that it contains eleven repeats and recognizes both RNA structure and sequence. Here we report a crystal structure of Saccharomyces cerevisiae Nop9 in complex with its target RNA within the 20S preribosomal RNA. This structure reveals that Nop9 brings together a carboxy-terminal module recognizing the 5' single-stranded region of the RNA and a bifunctional amino-terminal module recognizing the central double-stranded stem region. We further show that the 3' single-stranded region of the 20S target RNA adds sequence-independent binding energy to the RNA-Nop9 interaction. Both the amino- and carboxy-terminal modules retain the characteristic sequence-specific recognition of PUF proteins, but the amino-terminal module has also evolved a distinct interface, which allows Nop9 to recognize either single-stranded RNA sequences or RNAs with a combination of single-stranded and structured elements.