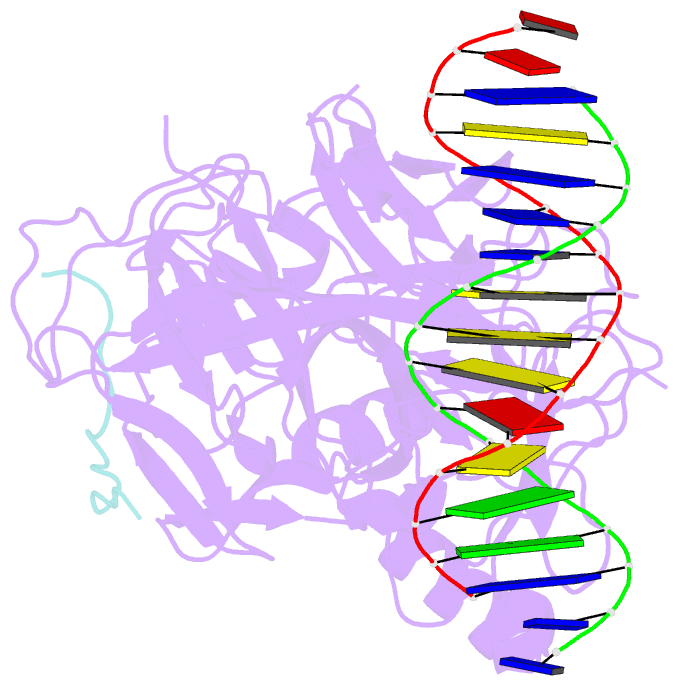
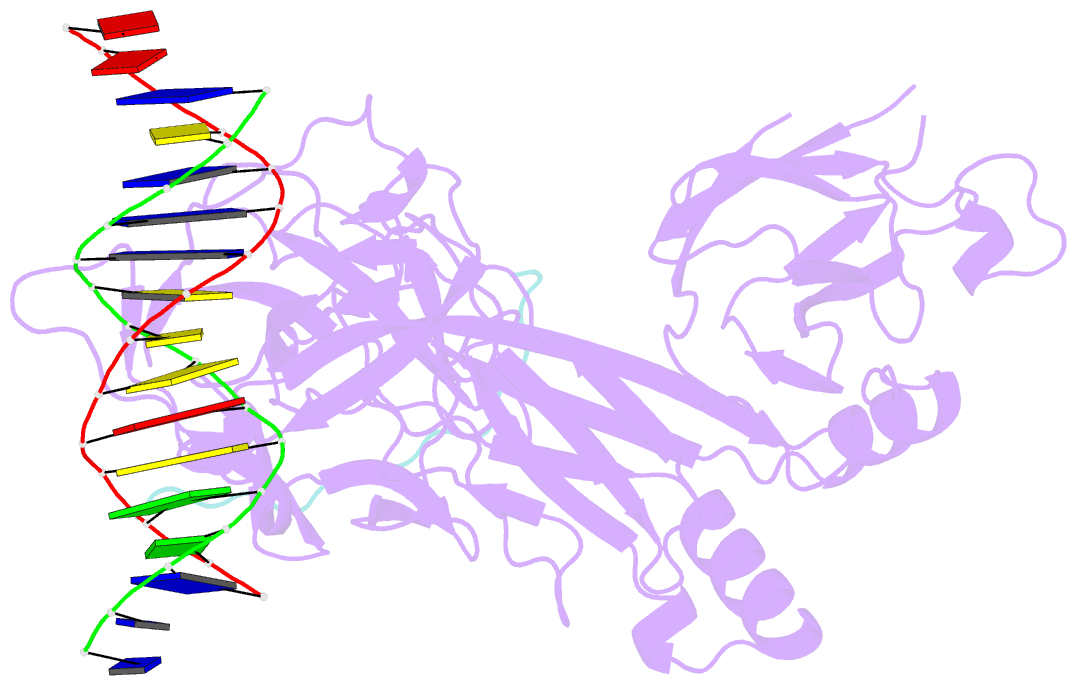
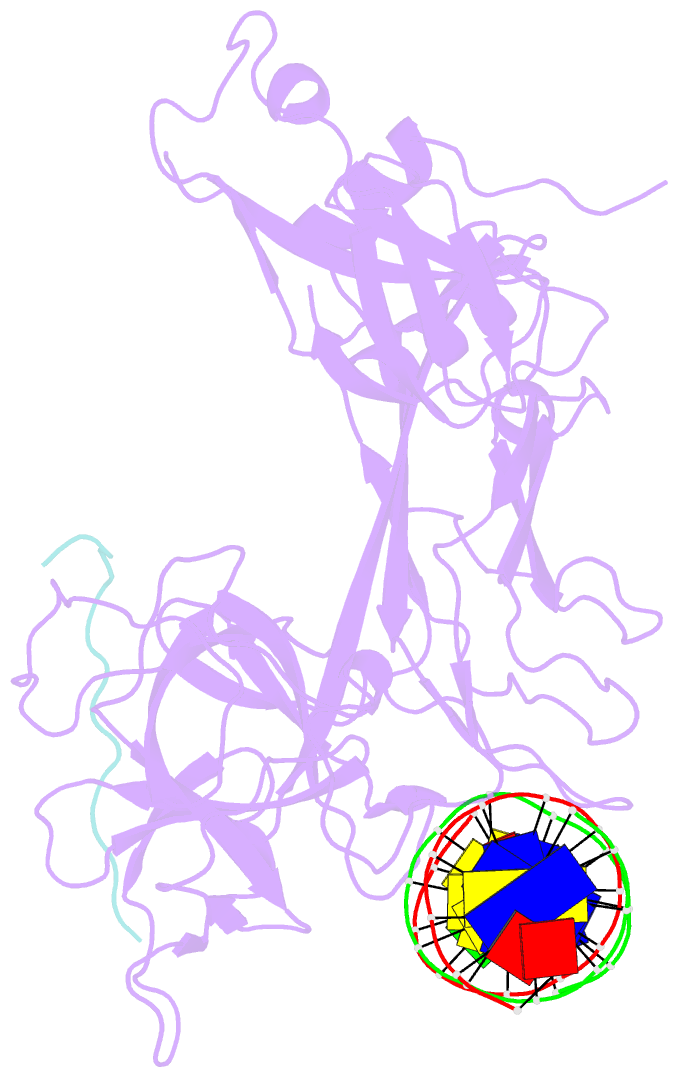
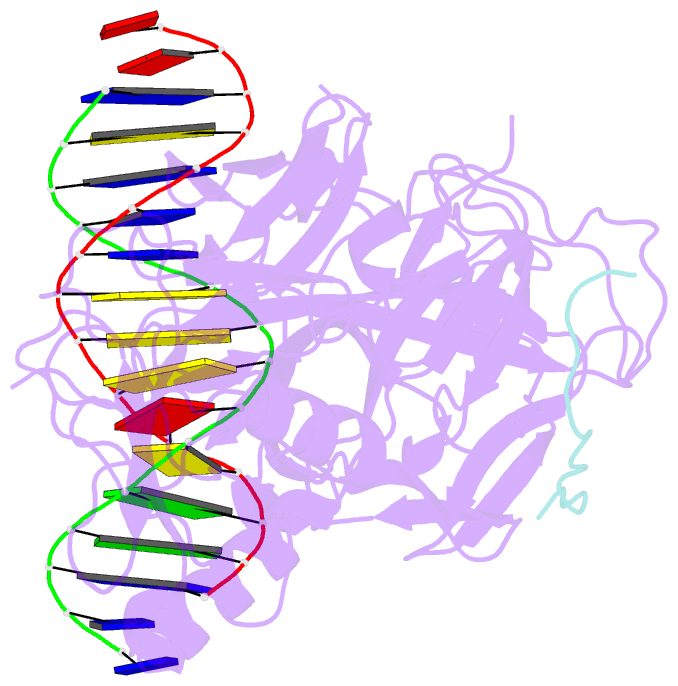
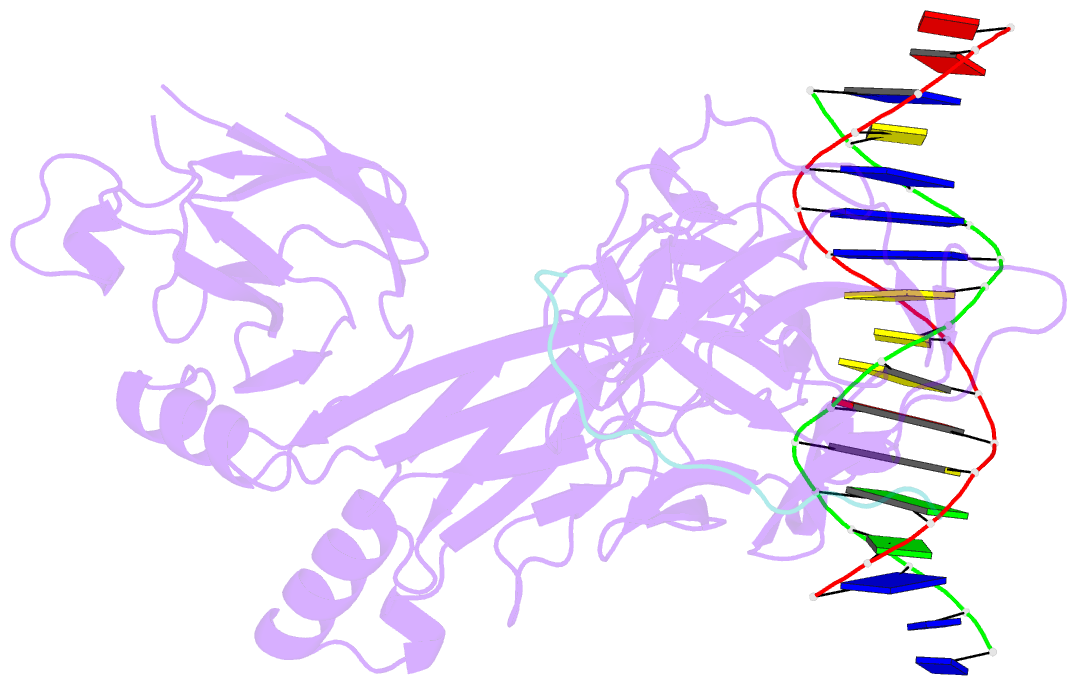
Summary information and primary citation
- PDB-id
- 6wqu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-signaling protein-DNA
- Method
- X-ray (2.41 Å)
- Summary
- Csl (rbpj) bound to notch3 ram and DNA
- Reference
- Landor SKJ, Santio NM, Eccleshall WB, Paramonov VM, Gagliani EK, Hall D, Jin SB, Dahlstrom KM, Salminen TA, Rivero-Muller A, Lendahl U, Kovall RA, Koskinen PJ, Sahlgren C (2021): "PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion." J.Biol.Chem., 296, 100593. doi: 10.1016/j.jbc.2021.100593.
- Abstract
- Dysregulation of the developmentally important Notch signaling pathway is implicated in several types of cancer, including breast cancer. However, the specific roles and regulation of the four different Notch receptors have remained elusive. We have previously reported that the oncogenic PIM kinases phosphorylate Notch1 and Notch3. Phosphorylation of Notch1 within the second nuclear localization sequence of its intracellular domain (ICD) enhances its transcriptional activity and tumorigenicity. In this study, we analyzed Notch3 phosphorylation and its functional impact. Unexpectedly, we observed that the PIM target sites are not conserved between Notch1 and Notch3. Notch3 ICD (N3ICD) is phosphorylated within a domain, which is essential for formation of a transcriptionally active complex with the DNA-binding protein CSL. Through molecular modeling, X-ray crystallography, and isothermal titration calorimetry, we demonstrate that phosphorylation of N3ICD sterically hinders its interaction with CSL and thereby inhibits its CSL-dependent transcriptional activity. Surprisingly however, phosphorylated N3ICD still maintains tumorigenic potential in breast cancer cells under estrogenic conditions, which support PIM expression. Taken together, our data indicate that PIM kinases modulate the signaling output of different Notch paralogs by targeting distinct protein domains and thereby promote breast cancer tumorigenesis via both CSL-dependent and CSL-independent mechanisms.