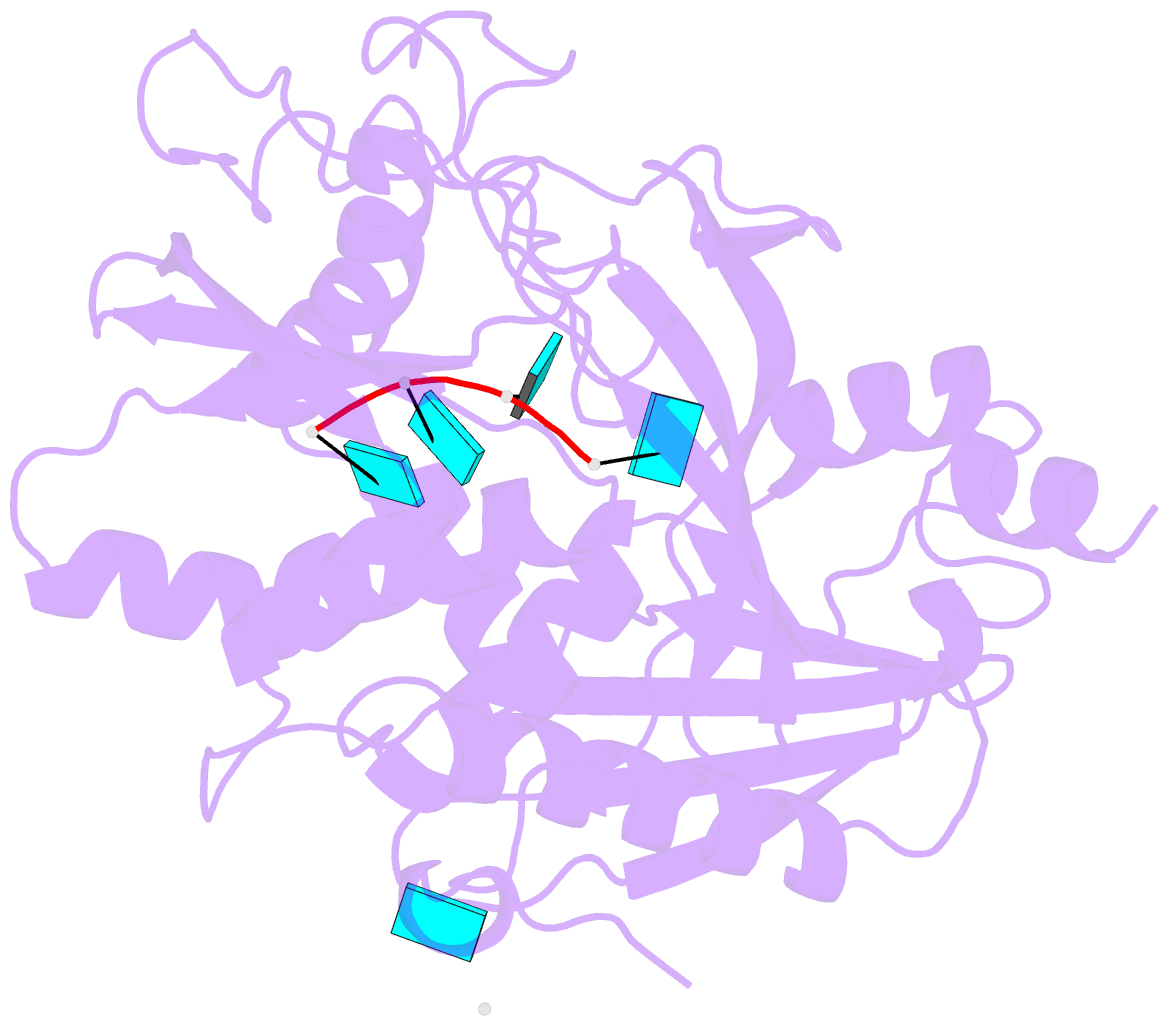
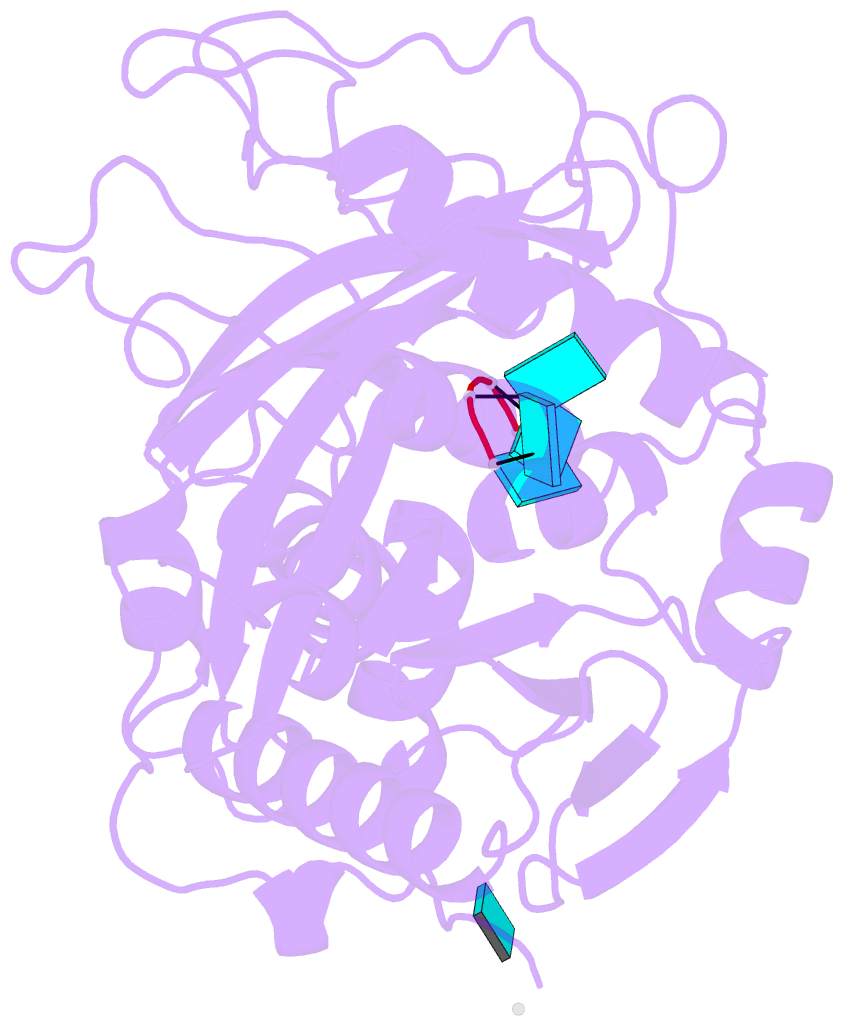
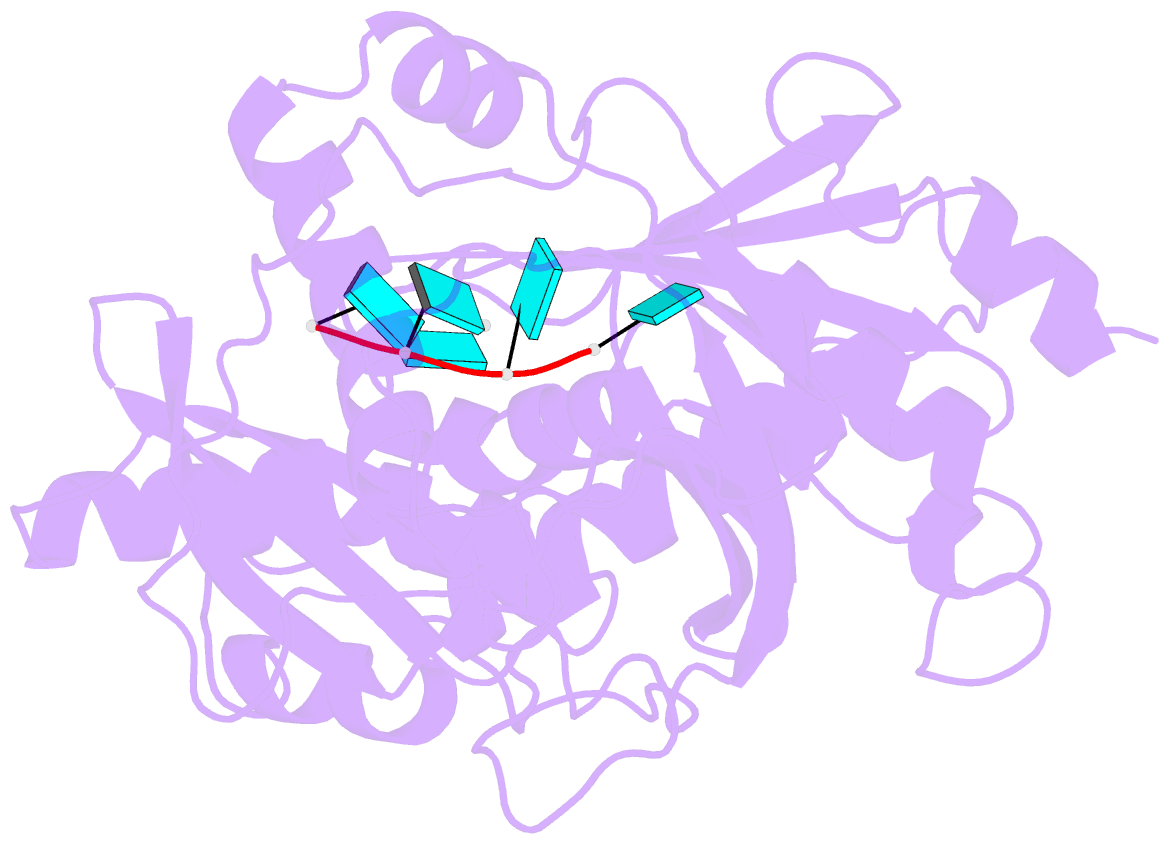
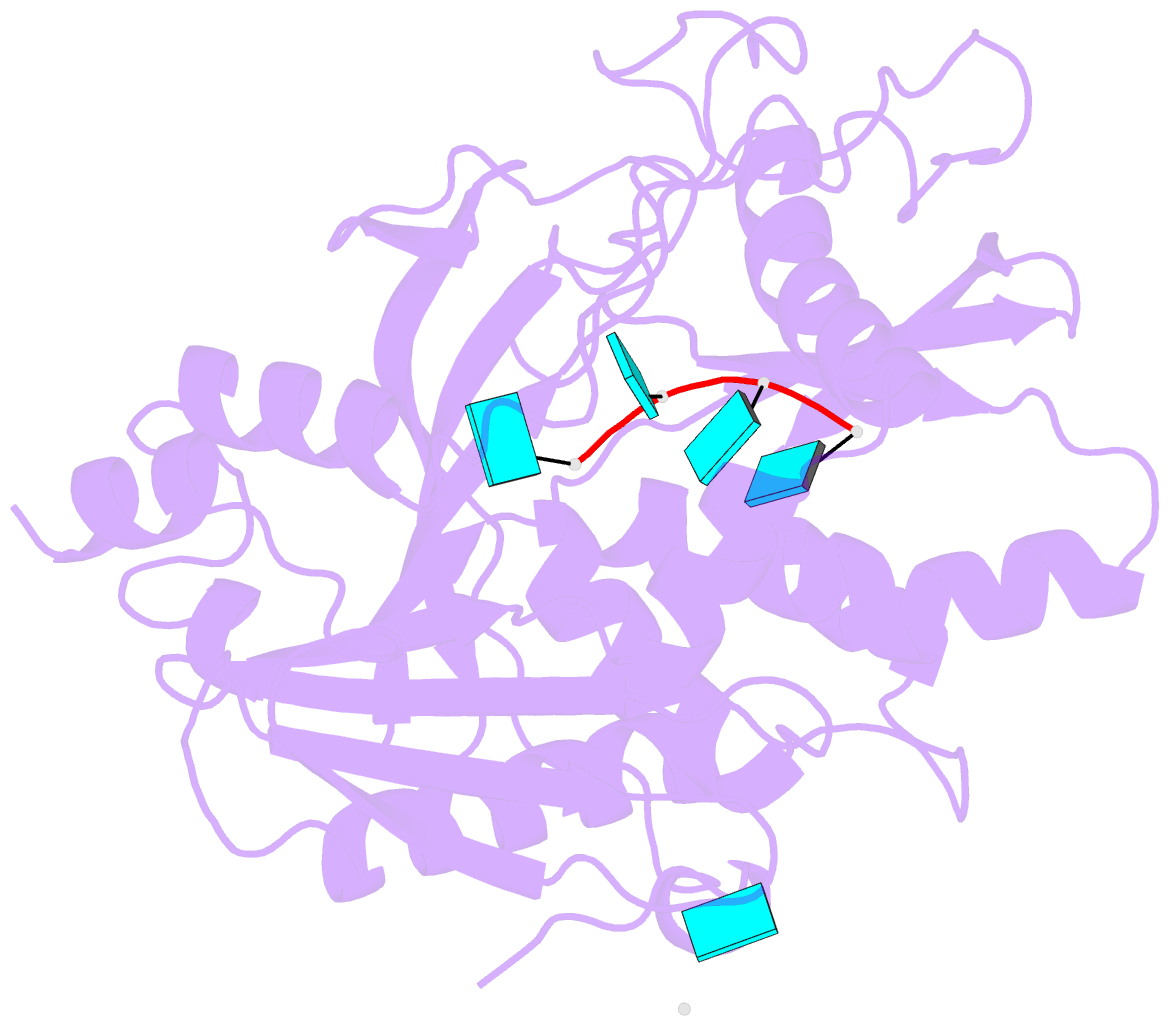
Summary information and primary citation
- PDB-id
- 6wre; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.0 Å)
- Summary
- Crystal structure of mouse dxo in complex with 5'-oh RNA substrate mimic and calcium ion
- Reference
- Doamekpor SK, Gozdek A, Kwasnik A, Kufel J, Tong L (2020): "A novel 5'-hydroxyl dinucleotide hydrolase activity for the DXO/Rai1 family of enzymes." Nucleic Acids Res., 48, 349-358. doi: 10.1093/nar/gkz1107.
- Abstract
- Modifications at the 5'-end of RNAs play a pivotal role in determining their fate. In eukaryotes, the DXO/Rai1 family of enzymes removes numerous 5'-end RNA modifications, thereby regulating RNA turnover. Mouse DXO catalyzes the elimination of incomplete 5'-end caps (including pyrophosphate) and the non-canonical NAD+ cap on mRNAs, and possesses distributive 5'-3' exoribonuclease activity toward 5'-monophosphate (5'-PO4) RNA. Here, we demonstrate that DXO also catalyzes the hydrolysis of RNAs bearing a 5'-hydroxyl group (5'-OH RNA). The crystal structure of DXO in complex with a 5'-OH RNA substrate mimic at 2.0 Å resolution provides elegant insight into the molecular mechanism of this activity. More importantly, the structure predicts that DXO first removes a dinucleotide from 5'-OH RNA. Our nuclease assays confirm this prediction and demonstrate that this 5'-hydroxyl dinucleotide hydrolase (HDH) activity for DXO is higher than the subsequent 5'-3' exoribonuclease activity for selected substrates. Fission yeast Rai1 also has HDH activity although it does not have 5'-3' exonuclease activity, and the Rat1-Rai1 complex can completely degrade 5'-OH RNA. An Arabidopsis DXO1 variant is active toward 5'-OH RNA but prefers 5'-PO4 RNA. Collectively, these studies demonstrate the diverse activities of DXO/Rai1 and expands the collection of RNA substrates that can undergo 5'-3' mediated decay.