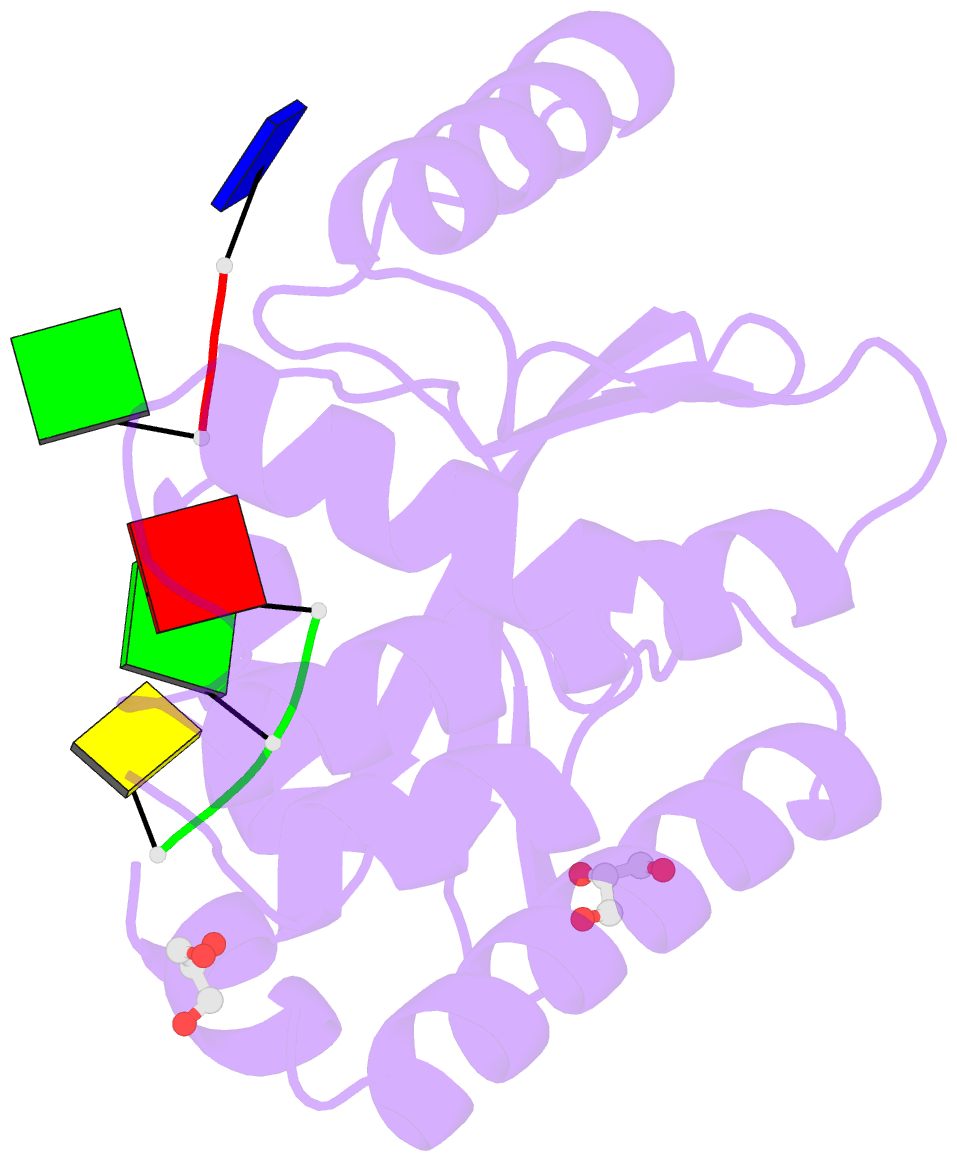
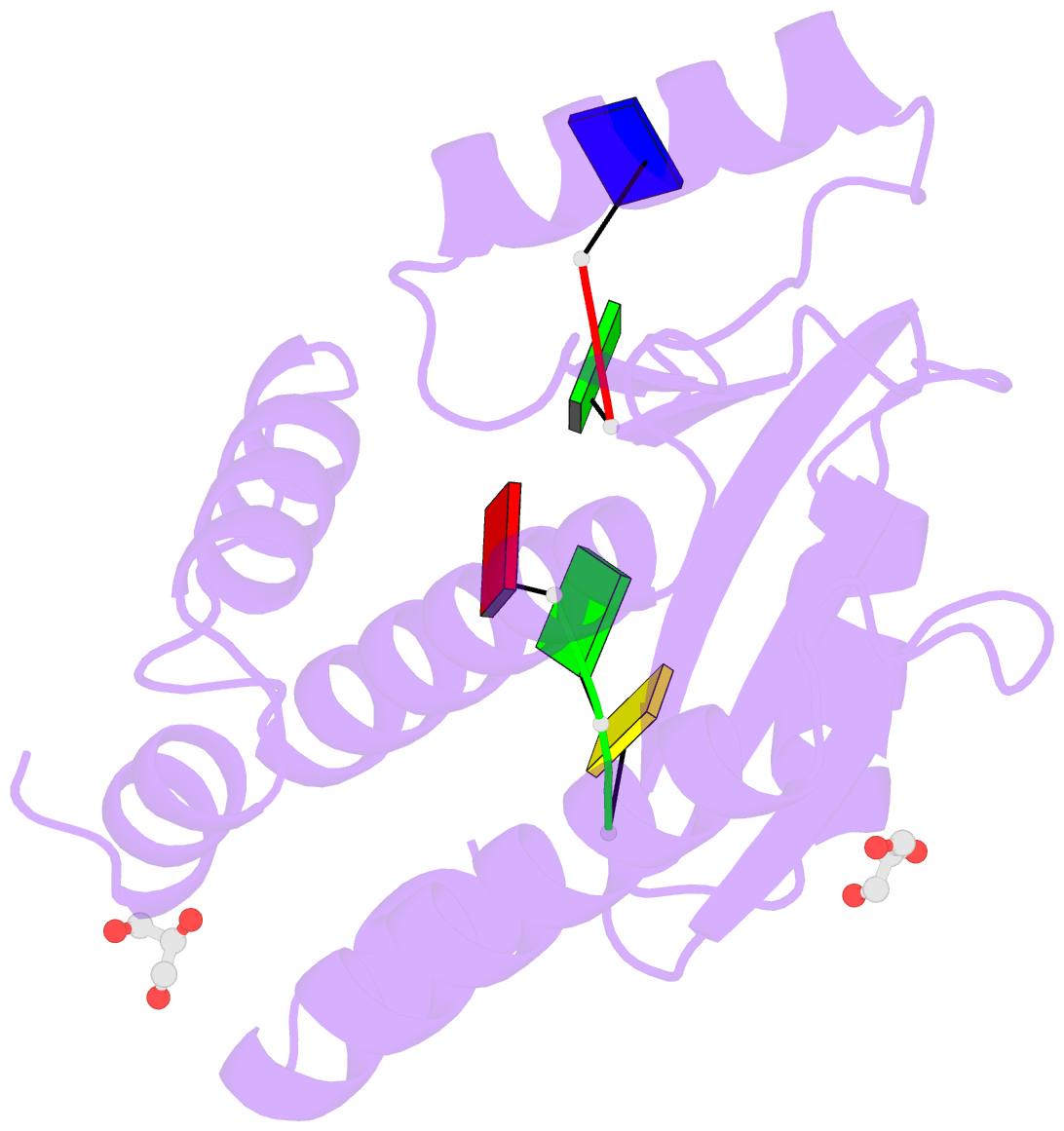
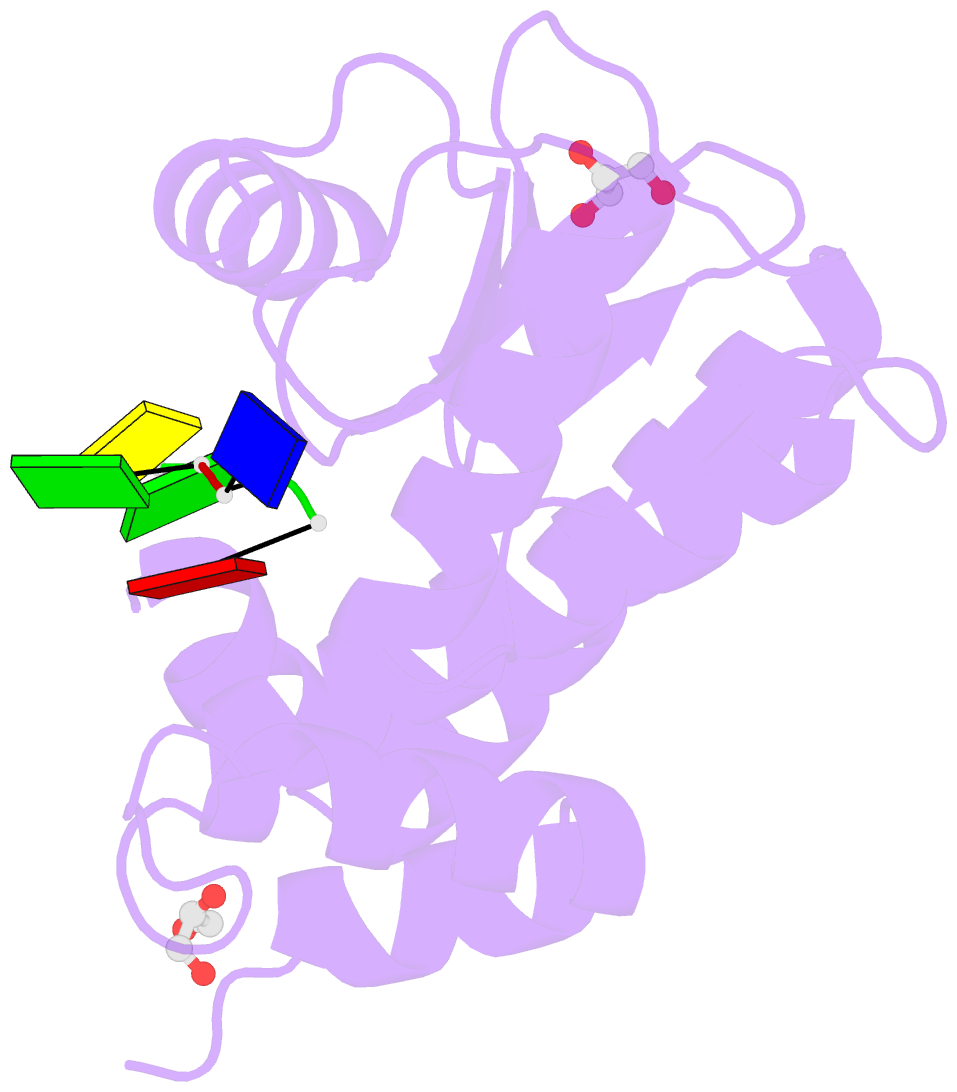
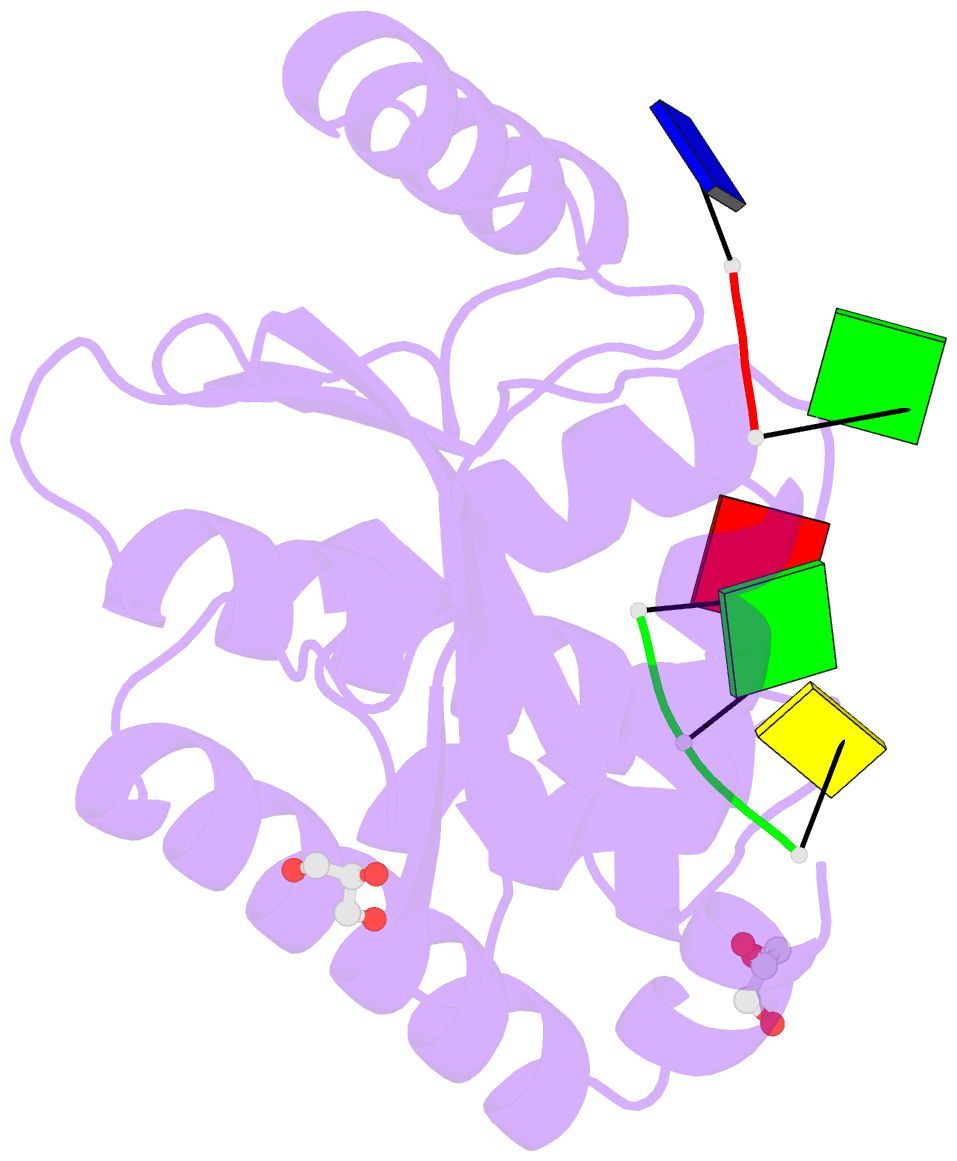
Summary information and primary citation
- PDB-id
- 6ws3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- X-ray (2.2 Å)
- Summary
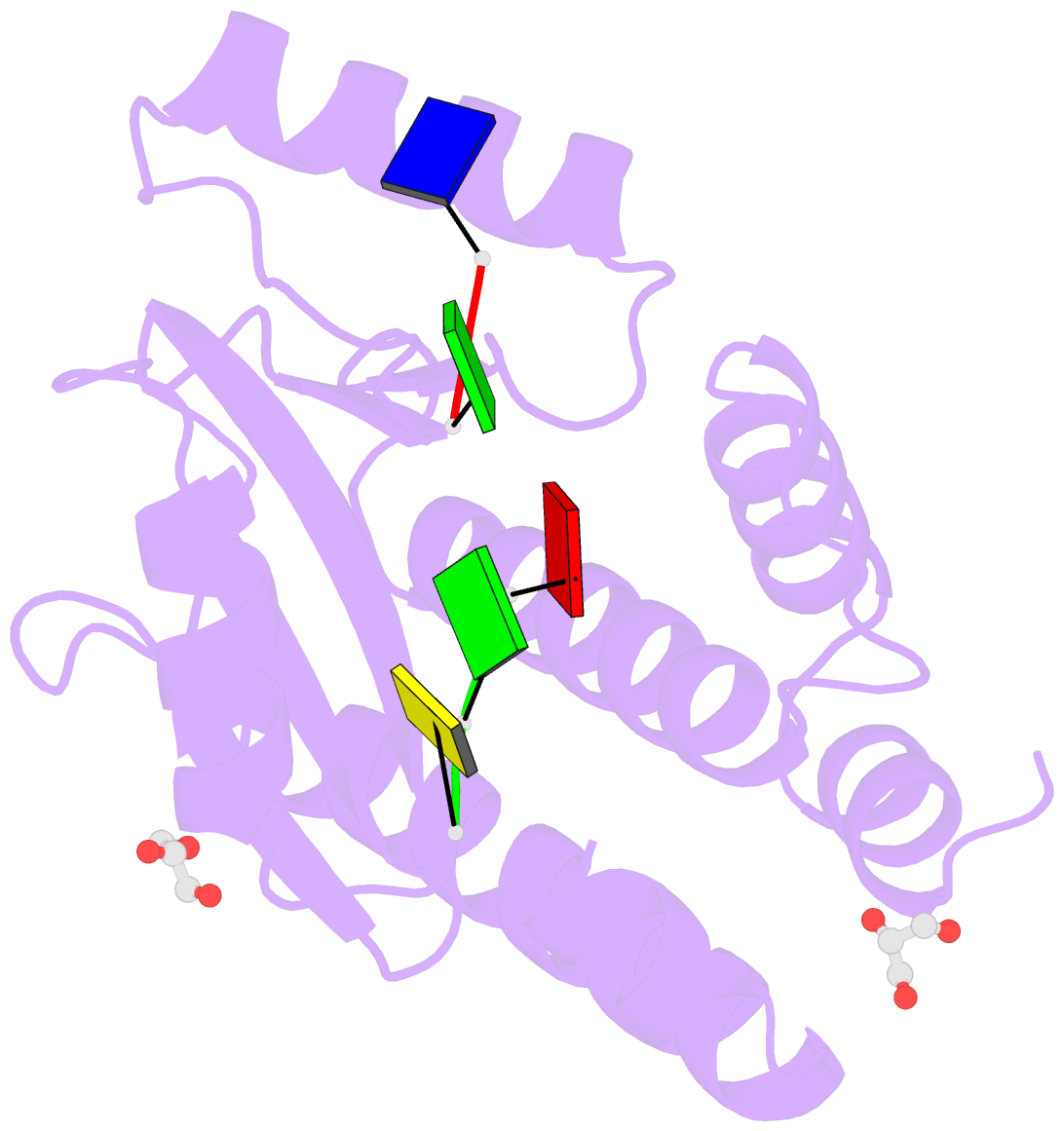
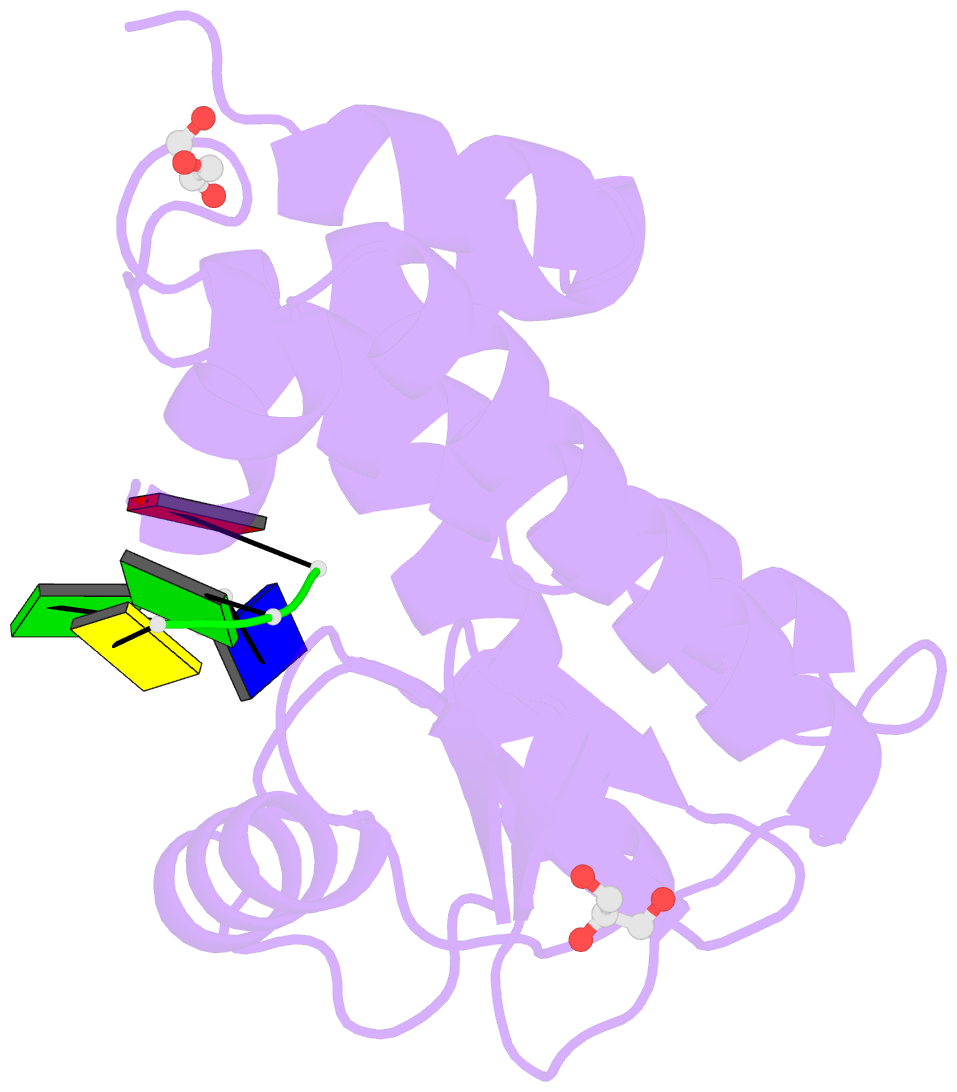
- The crystal structure of the 2009-h1n1-california pa endonuclease wild type bound to DNA oligomers tg and agca (from cleaved gtgagcagtg)
- Reference
- Kumar G, Cuypers M, Webby RR, Webb TR, White SW (2021): "Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism." Nucleic Acids Res., 49, 1609-1618. doi: 10.1093/nar/gkaa1294.
- Abstract
- The endonuclease activity within the influenza virus cap-snatching process is a proven therapeutic target. The anti-influenza drug baloxavir is highly effective, but is associated with resistance mutations that threaten its clinical efficacy. The endonuclease resides within the N-terminal domain of the PA subunit (PAN) of the influenza RNA dependent RNA polymerase, and we report here complexes of PAN with RNA and DNA oligonucleotides to understand its specificity and the structural basis of baloxavir resistance mutations. The RNA and DNA oligonucleotides bind within the substrate binding groove of PAN in a similar fashion, explaining the ability of the enzyme to cleave both substrates. The individual nucleotides occupy adjacent conserved pockets that flank the two-metal active site. However, the 2' OH of the RNA ribose moieties engage in additional interactions that appear to optimize the binding and cleavage efficiency for the natural substrate. The major baloxavir resistance mutation at position 38 is at the core of the substrate binding site, but structural studies and modeling suggest that it maintains the necessary virus fitness via compensating interactions with RNA. These studies will facilitate the development of new influenza therapeutics that spatially match the substrate and are less likely to elicit resistance mutations.