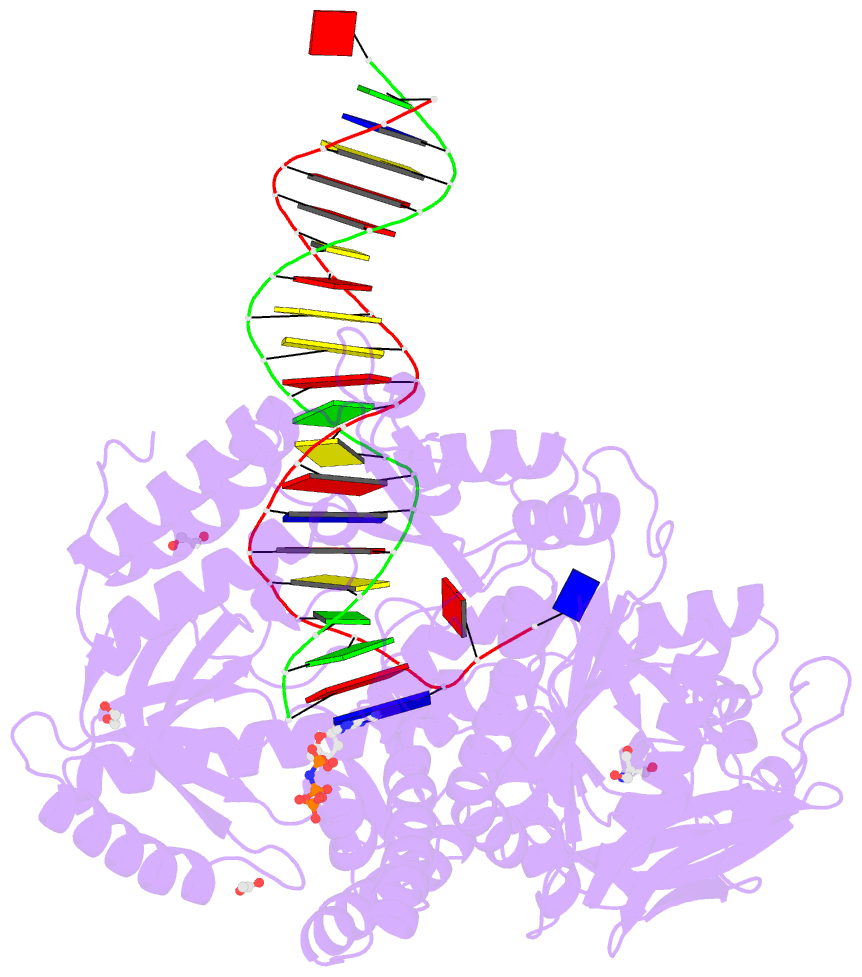
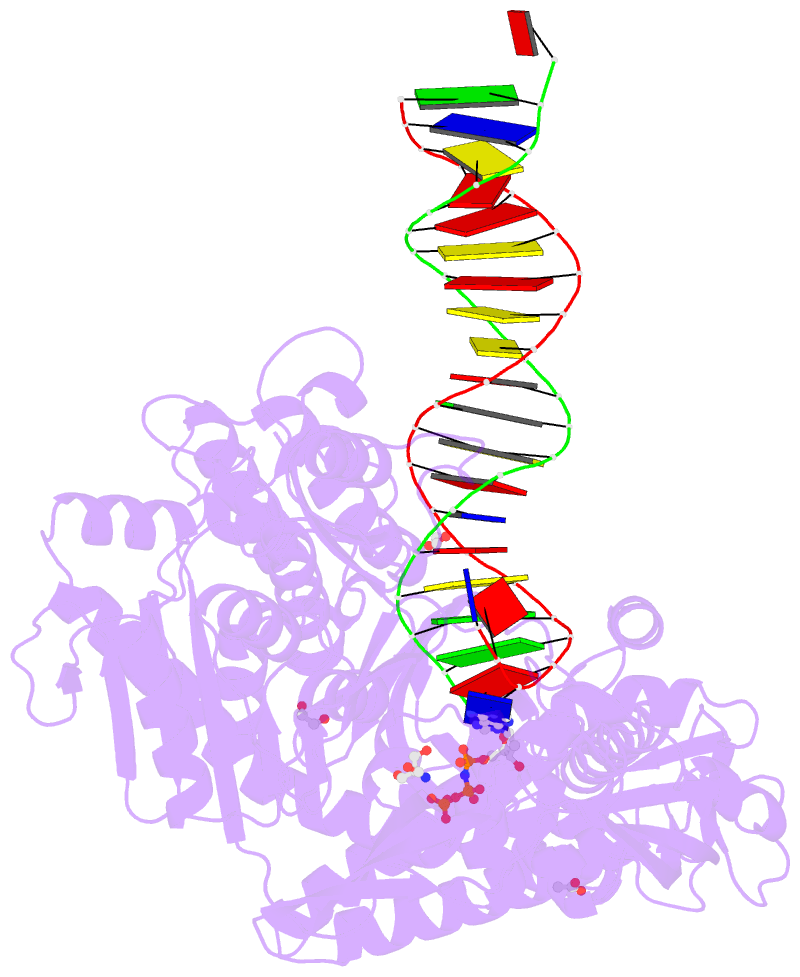
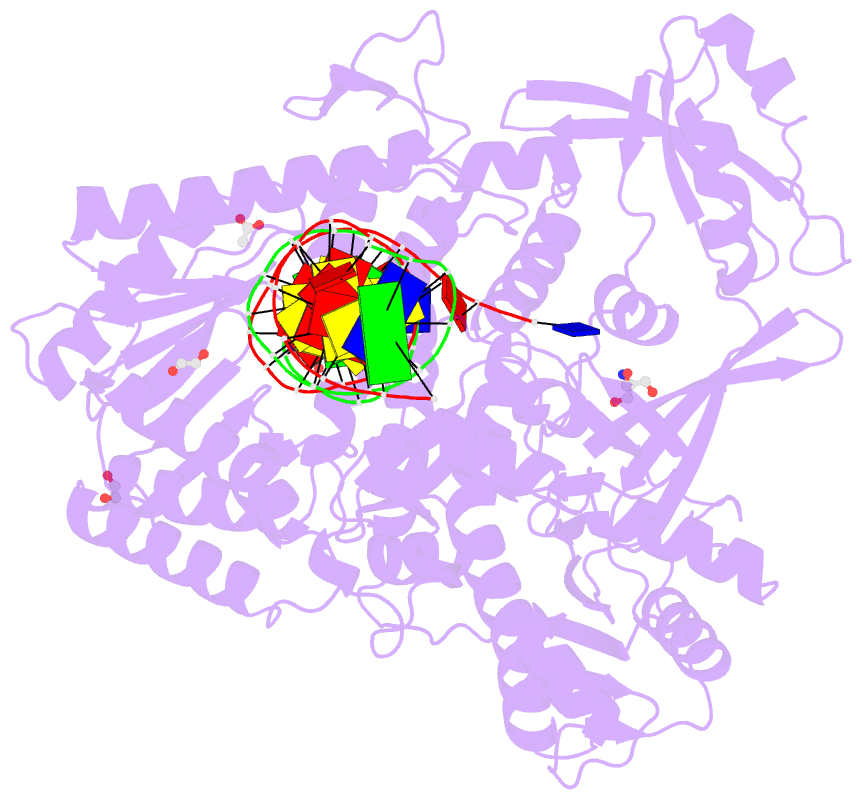
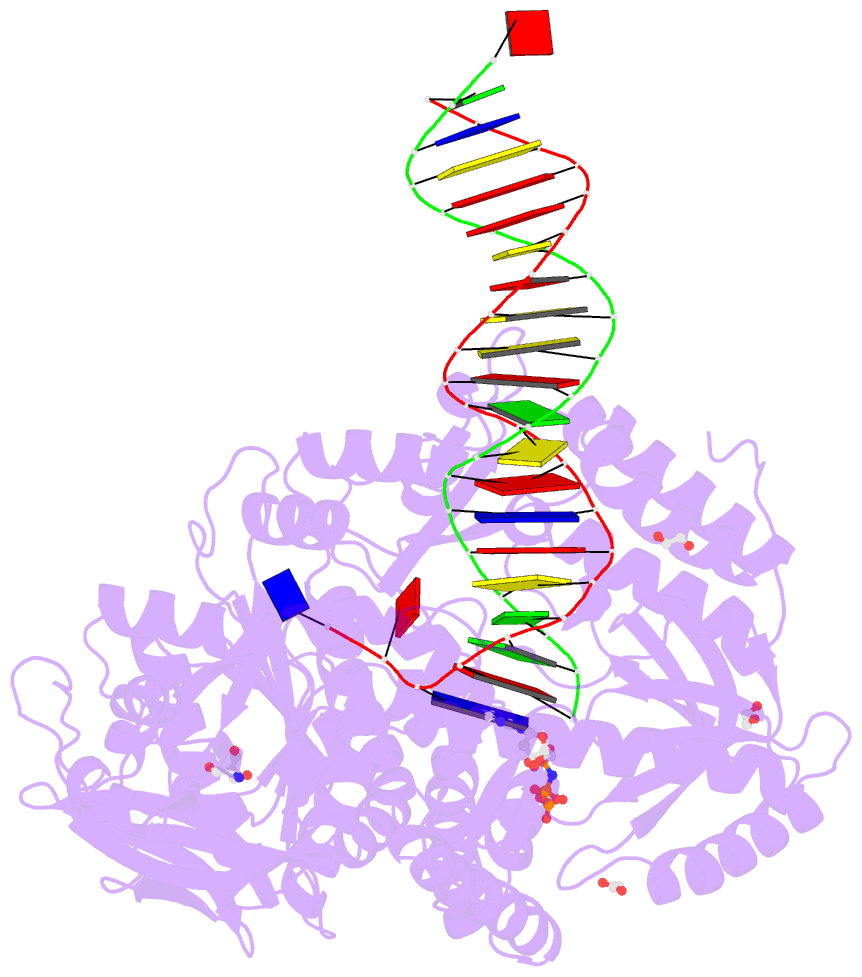
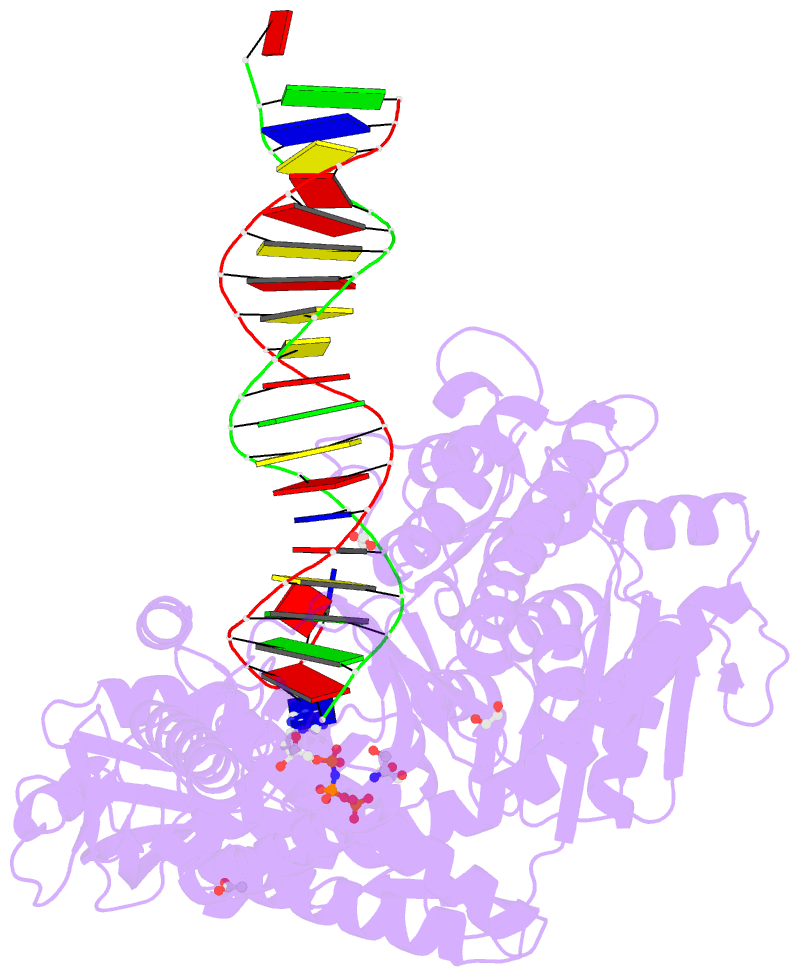
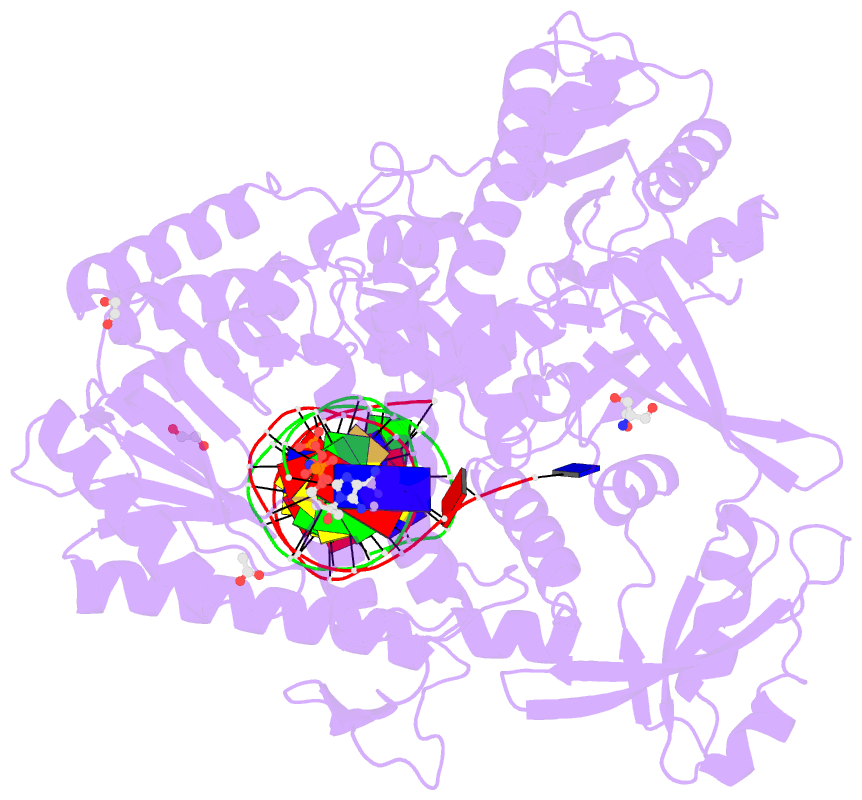
Summary information and primary citation
- PDB-id
- 6wya; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.41 Å)
- Summary
- Rtx (reverse transcription xenopolymerase) in complex with a DNA duplex and dampnpp
- Reference
- Choi WS, He P, Pothukuchy A, Gollihar J, Ellington AD, Yang W (2020): "How a B family DNA polymerase has been evolved to copy RNA." Proc.Natl.Acad.Sci.USA, 117, 21274-21280. doi: 10.1073/pnas.2009415117.
- Abstract
- We report here crystal structures of a reverse transcriptase RTX, which was evolved in vitro from the B family polymerase KOD, in complex with either a DNA duplex or an RNA-DNA hybrid. Compared with the apo, binary, and ternary complex structures of the original KOD polymerase, the 16 substitutions that result in the function of copying RNA to DNA do not change the overall protein structure. Only six substitutions occur at the substrate-binding surface, and the others change domain-domain interfaces in the polymerase to enable RNA-DNA hybrid binding and reverse transcription. Most notably, F587L at the Palm and Thumb interface stabilizes the open and apo conformation of the Thumb. The intrinsically flexible Thumb domain seems to play a major role in accommodating the RNA-DNA hybrid product distal to the active site. This is reminiscent of naturally occurring RNA-dependent DNA polymerases, including telomerase, which have a dramatically augmented Thumb domain, and of reverse transcriptase, which extends its Thumb with the RNase H domain.