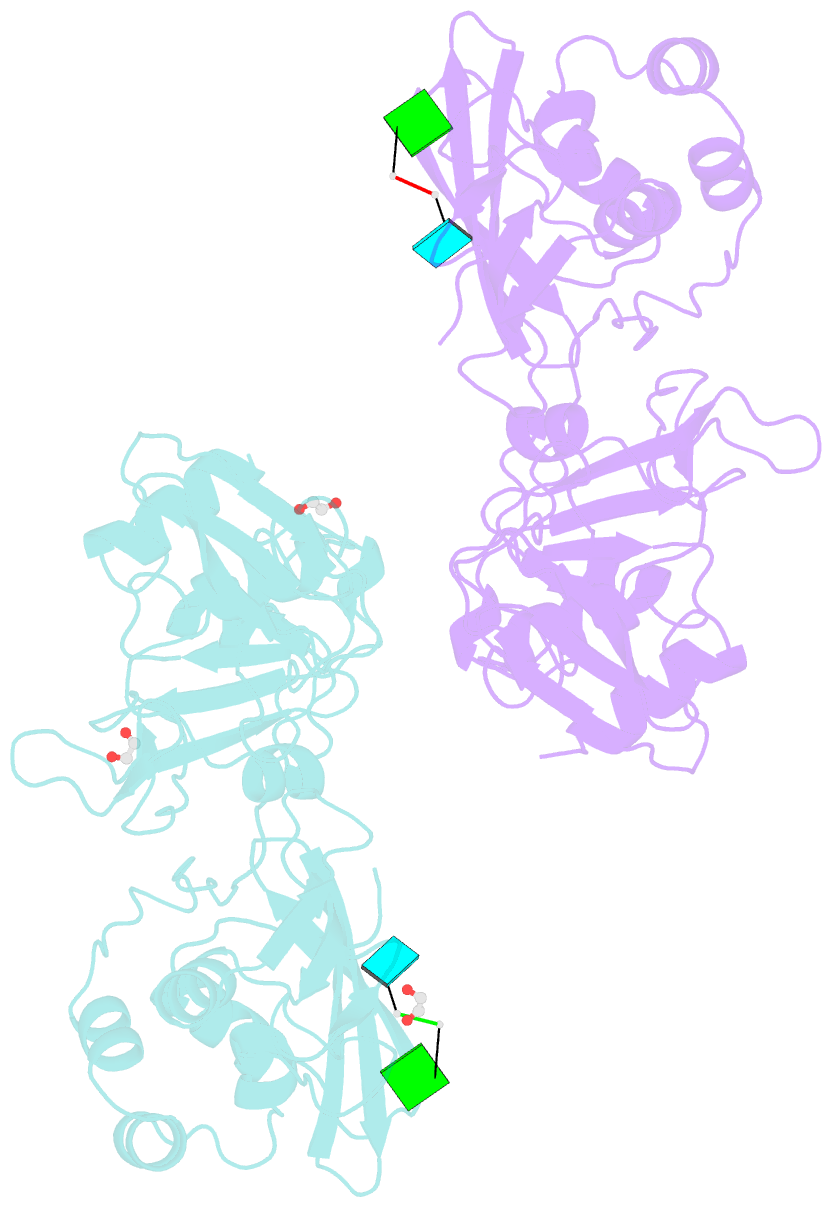
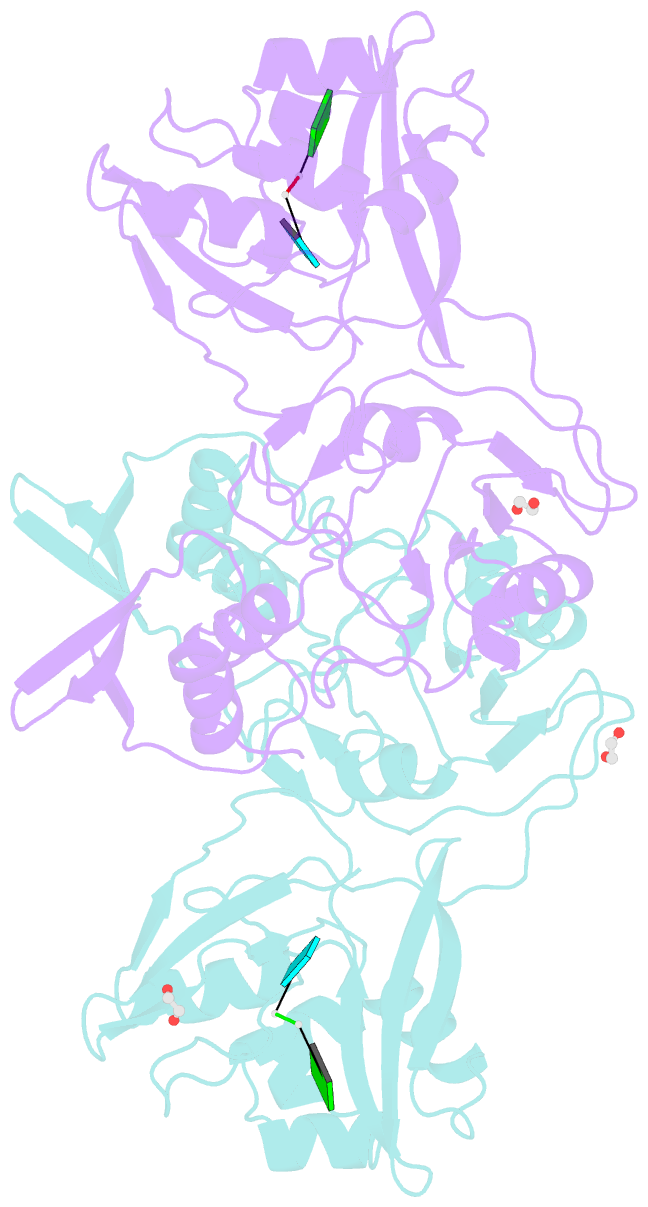
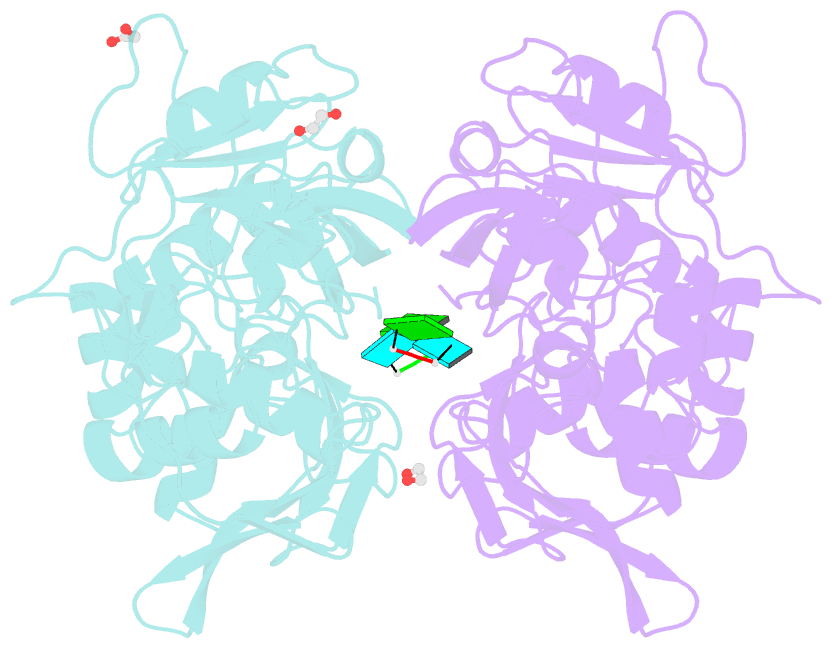
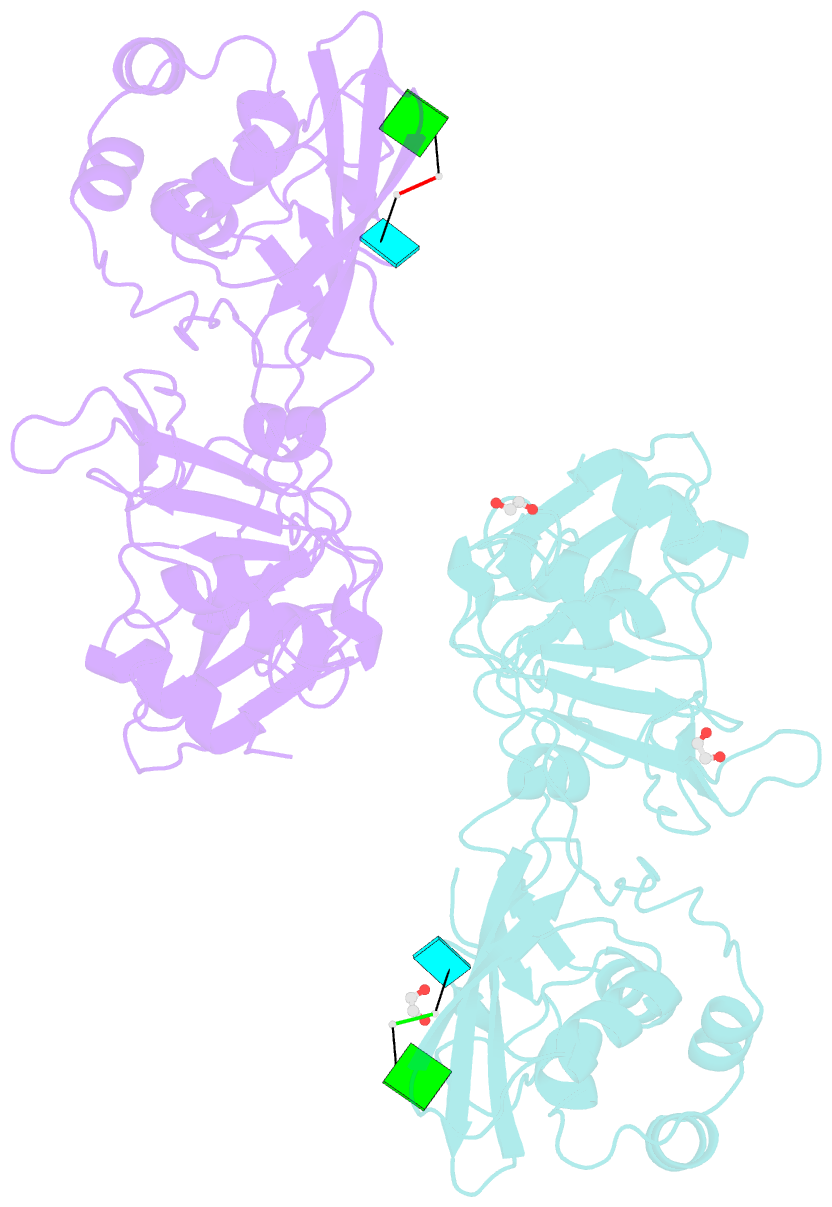
Summary information and primary citation
- PDB-id
- 6x1b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- X-ray (1.97 Å)
- Summary
- Crystal structure of nsp15 endoribonuclease from sars cov-2 in the complex with the product nucleotide gpu.
- Reference
- Kim Y, Wower J, Maltseva N, Chang C, Jedrzejczak R, Wilamowski M, Kang S, Nicolaescu V, Randall G, Michalska K, Joachimiak A (2021): "Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2." Commun Biol, 4, 193. doi: 10.1038/s42003-021-01735-9.
- Abstract
- SARS-CoV-2 Nsp15 is a uridine-specific endoribonuclease with C-terminal catalytic domain belonging to the EndoU family that is highly conserved in coronaviruses. As endoribonuclease activity seems to be responsible for the interference with the innate immune response, Nsp15 emerges as an attractive target for therapeutic intervention. Here we report the first structures with bound nucleotides and show how the enzyme specifically recognizes uridine moiety. In addition to a uridine site we present evidence for a second base binding site that can accommodate any base. The structure with a transition state analog, uridine vanadate, confirms interactions key to catalytic mechanisms. In the presence of manganese ions, the enzyme cleaves unpaired RNAs. This acquired knowledge was instrumental in identifying Tipiracil, an FDA approved drug that is used in the treatment of colorectal cancer, as a potential anti-COVID-19 drug. Using crystallography, biochemical, and whole-cell assays, we demonstrate that Tipiracil inhibits SARS-CoV-2 Nsp15 by interacting with the uridine binding pocket in the enzyme's active site. Our findings provide new insights for the development of uracil scaffold-based drugs.