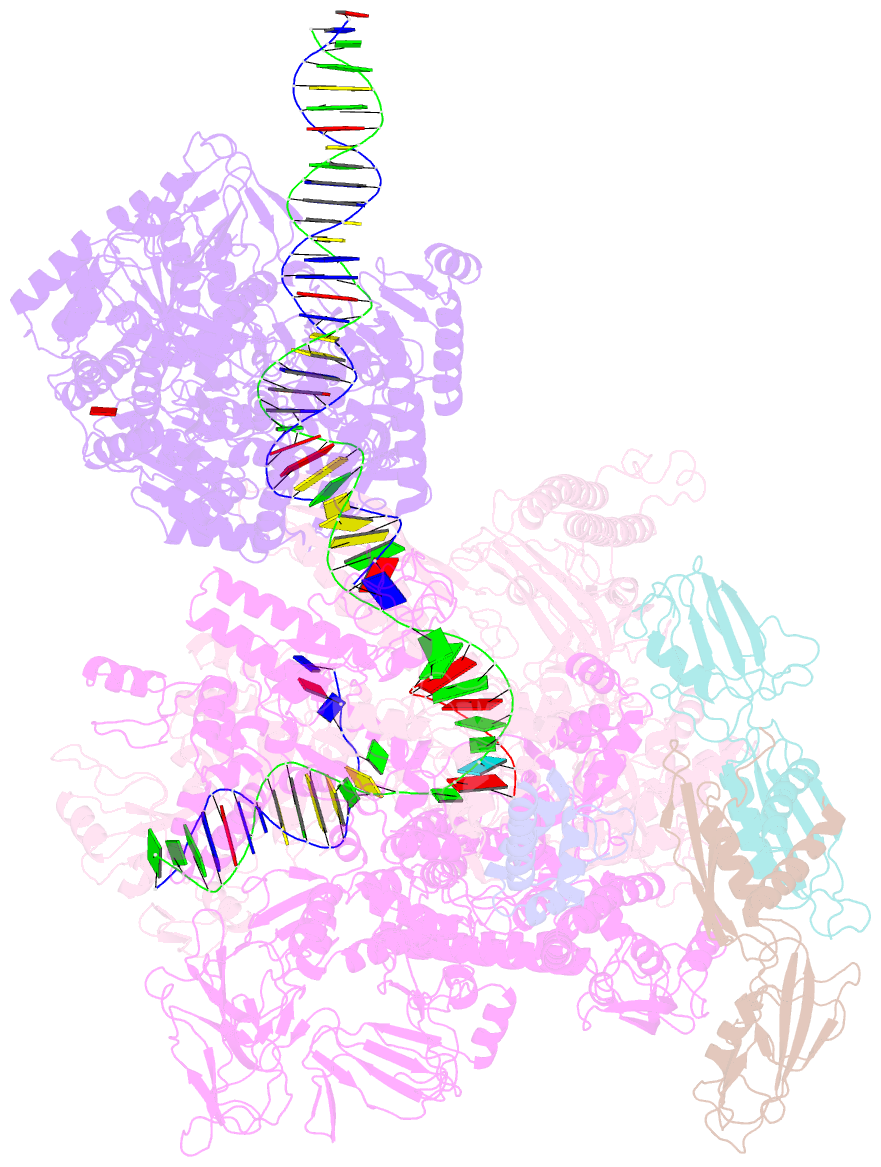
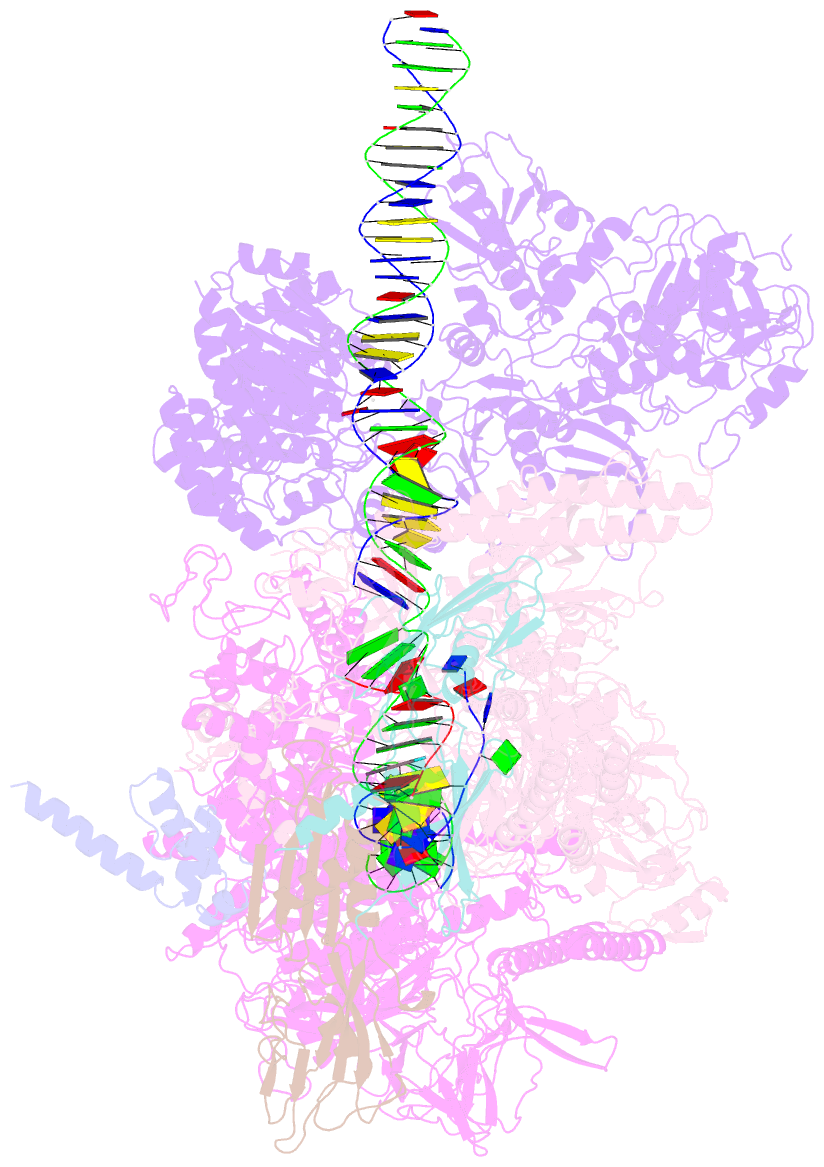
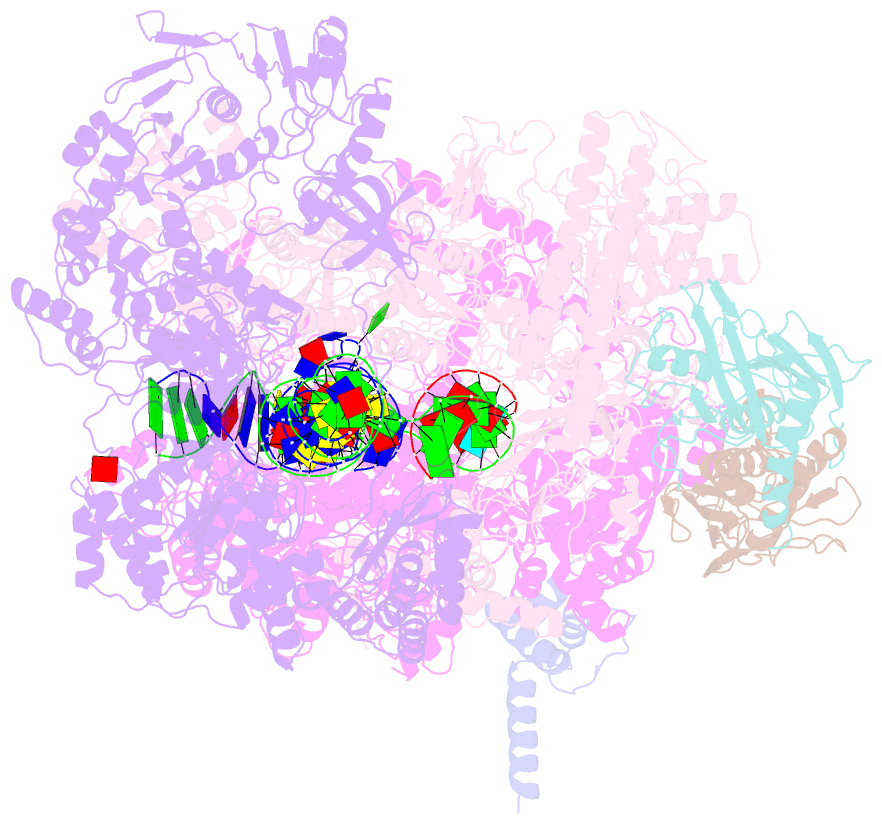
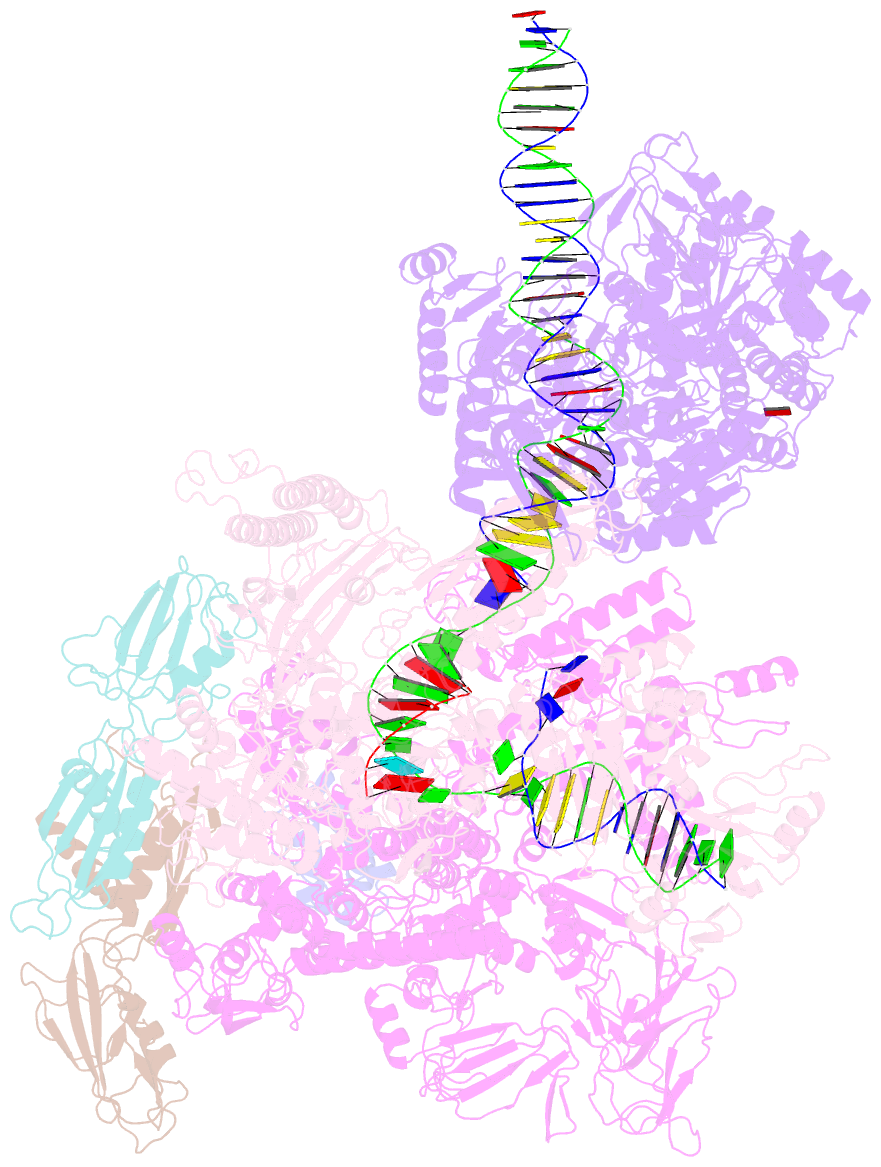
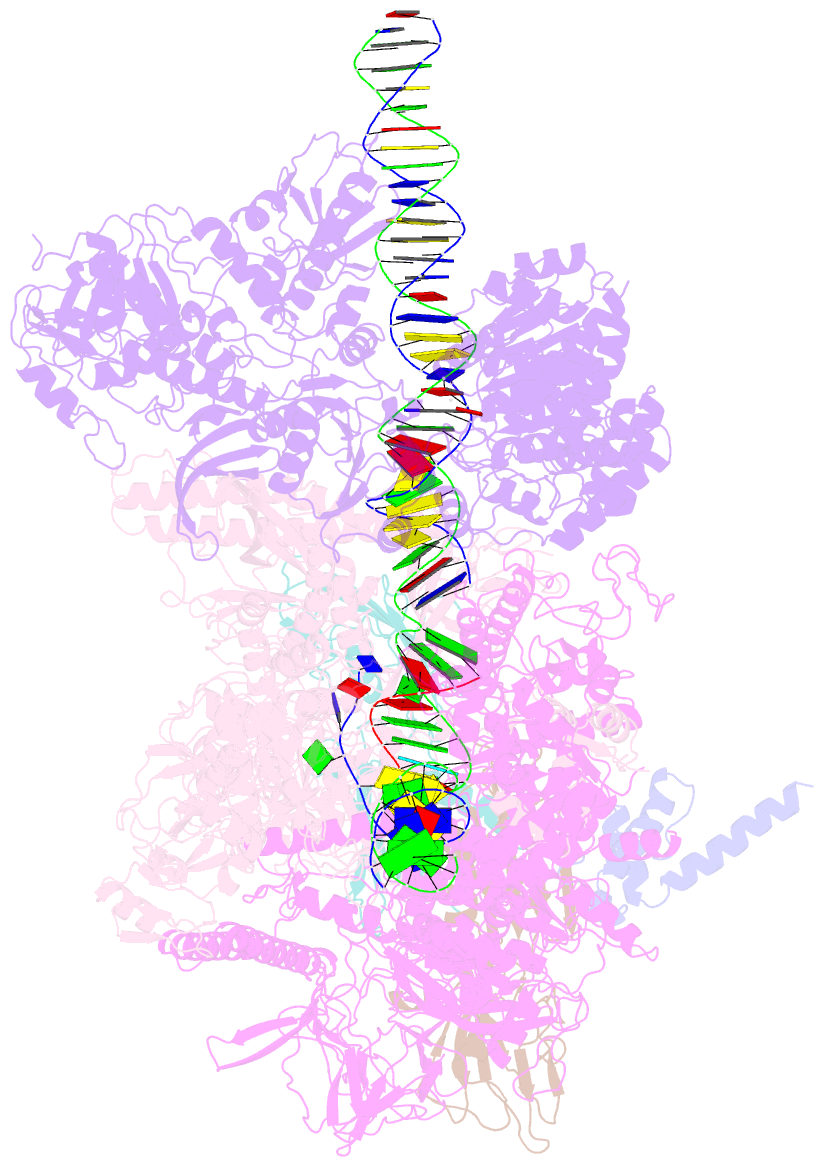
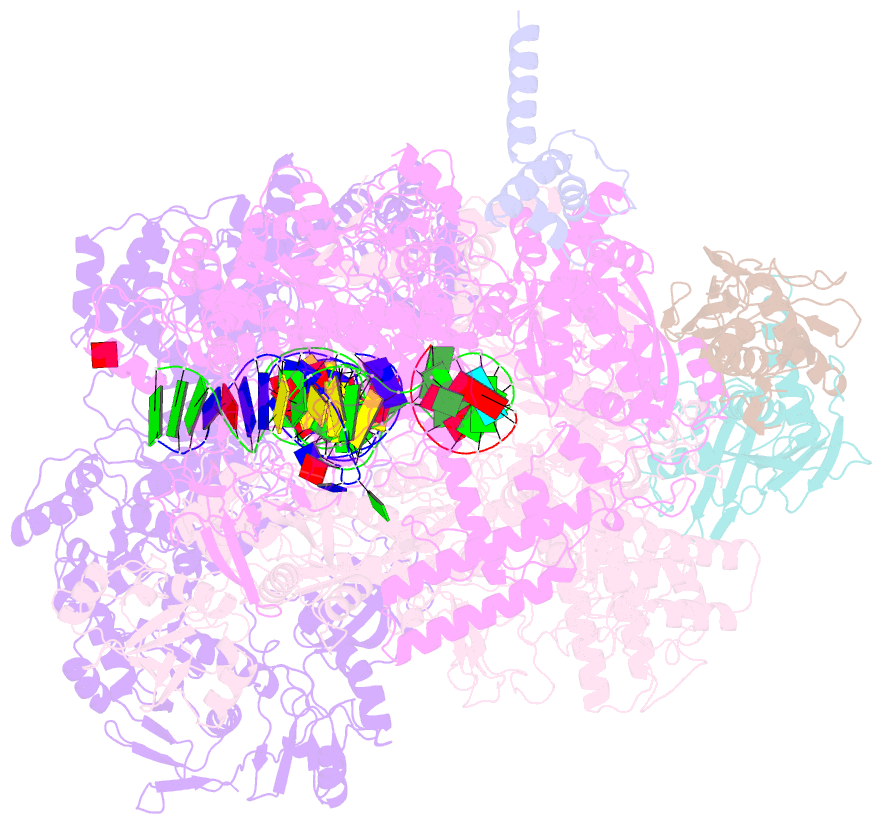
Summary information and primary citation
- PDB-id
- 6x2f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-RNA-DNA
- Method
- cryo-EM (4.0 Å)
- Summary
- Mfd-bound e.coli RNA polymerase elongation complex - l2 state
- Reference
- Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, Darst SA (2021): "Structural basis for transcription complex disruption by the Mfd translocase." Elife, 10. doi: 10.7554/eLife.62117.
- Abstract
- Transcription-coupled repair (TCR) is a sub-pathway of nucleotide excision repair (NER) that preferentially removes lesions from the template-strand (t-strand) that stall RNA polymerase (RNAP) elongation complexes (ECs). Mfd mediates TCR in bacteria by removing the stalled RNAP concealing the lesion and recruiting Uvr(A)BC. We used cryo-electron microscopy to visualize Mfd engaging with a stalled EC and attempting to dislodge the RNAP. We visualized seven distinct Mfd-EC complexes in both ATP and ADP-bound states. The structures explain how Mfd is remodeled from its repressed conformation, how the UvrA-interacting surface of Mfd is hidden during most of the remodeling process to prevent premature engagement with the NER pathway, how Mfd alters the RNAP conformation to facilitate disassembly, and how Mfd forms a processive translocation complex after dislodging the RNAP. Our results reveal an elaborate mechanism for how Mfd kinetically discriminates paused from stalled ECs and disassembles stalled ECs to initiate TCR.