Summary information and primary citation
- PDB-id
-
6xeo;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- DNA binding protein, hydrolase-DNA
- Method
- cryo-EM (5.5 Å)
- Summary
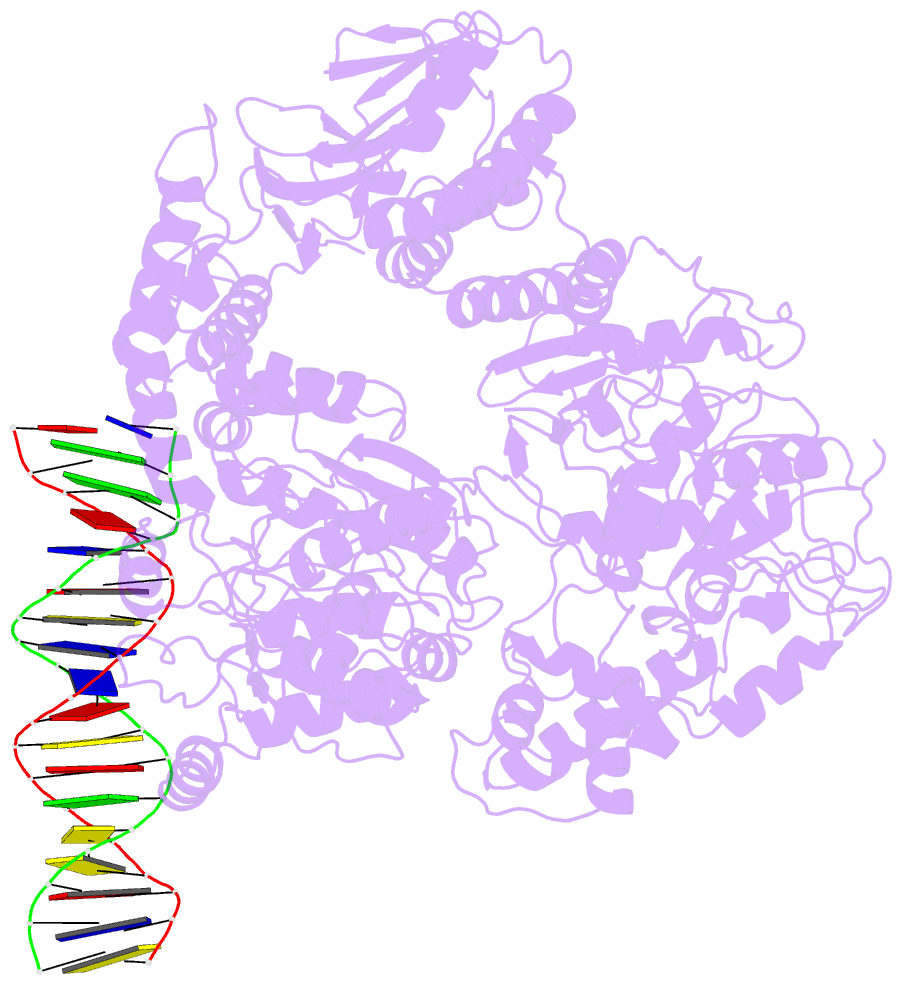
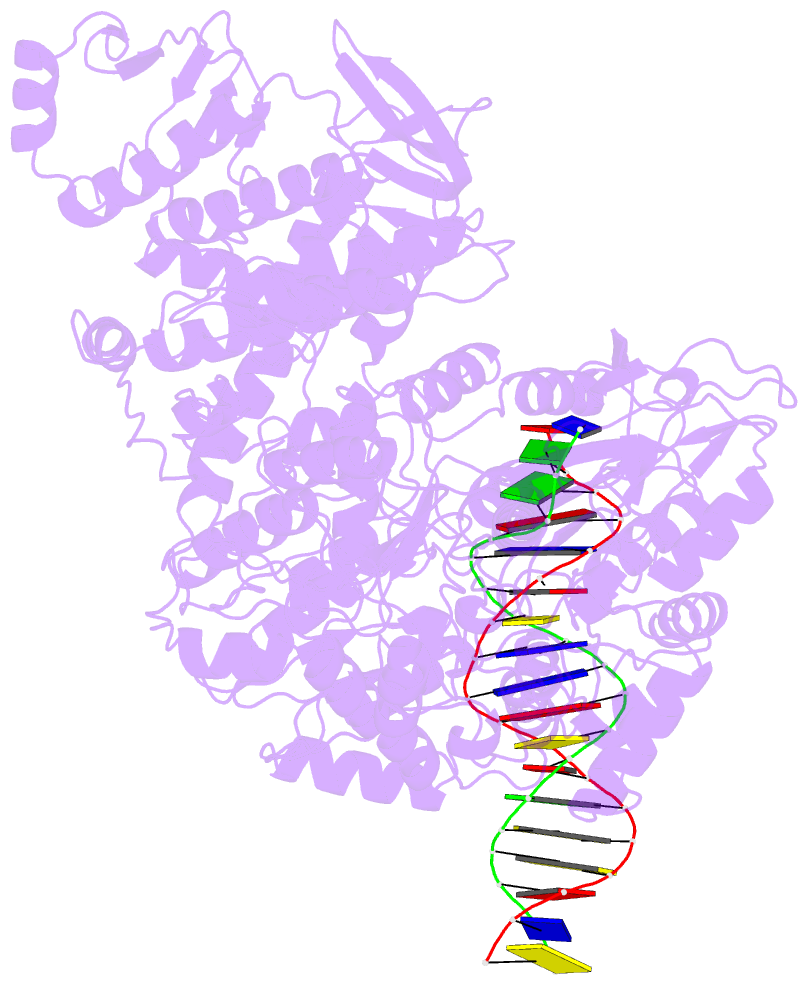
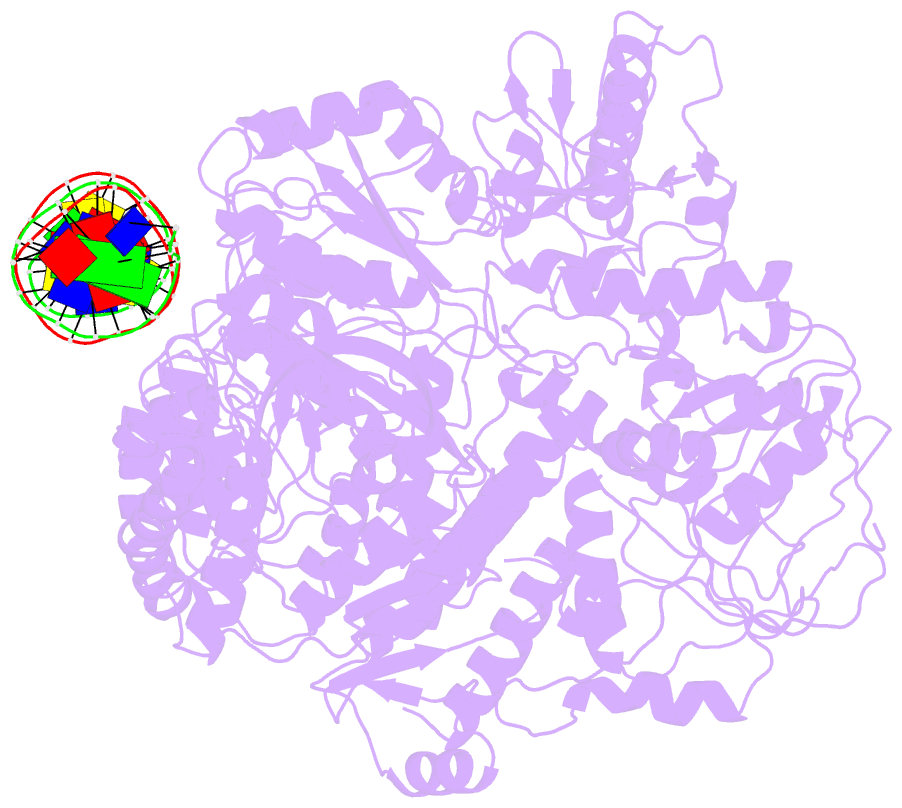
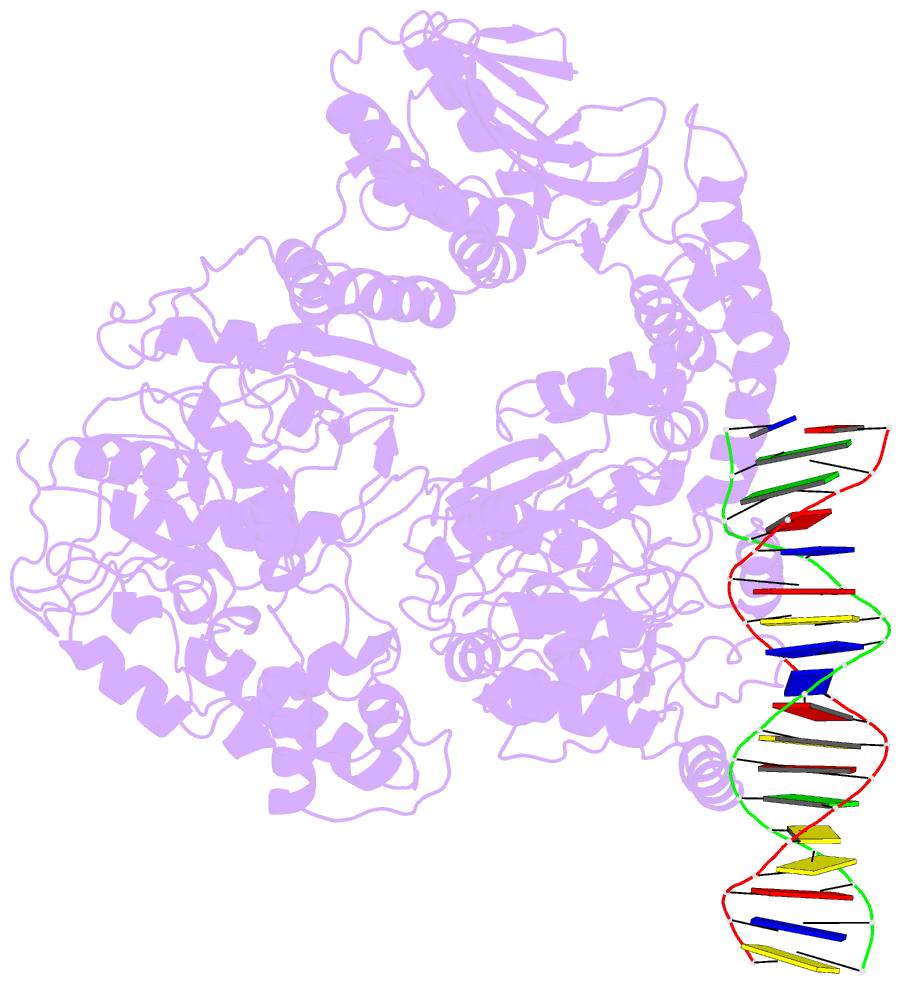
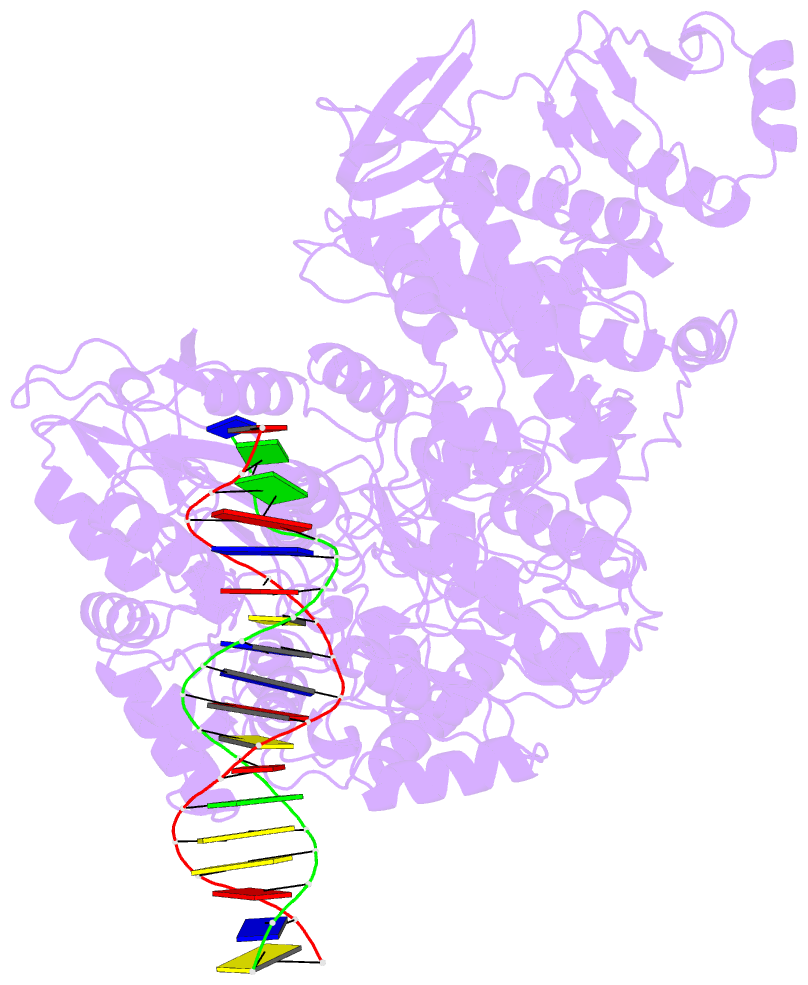
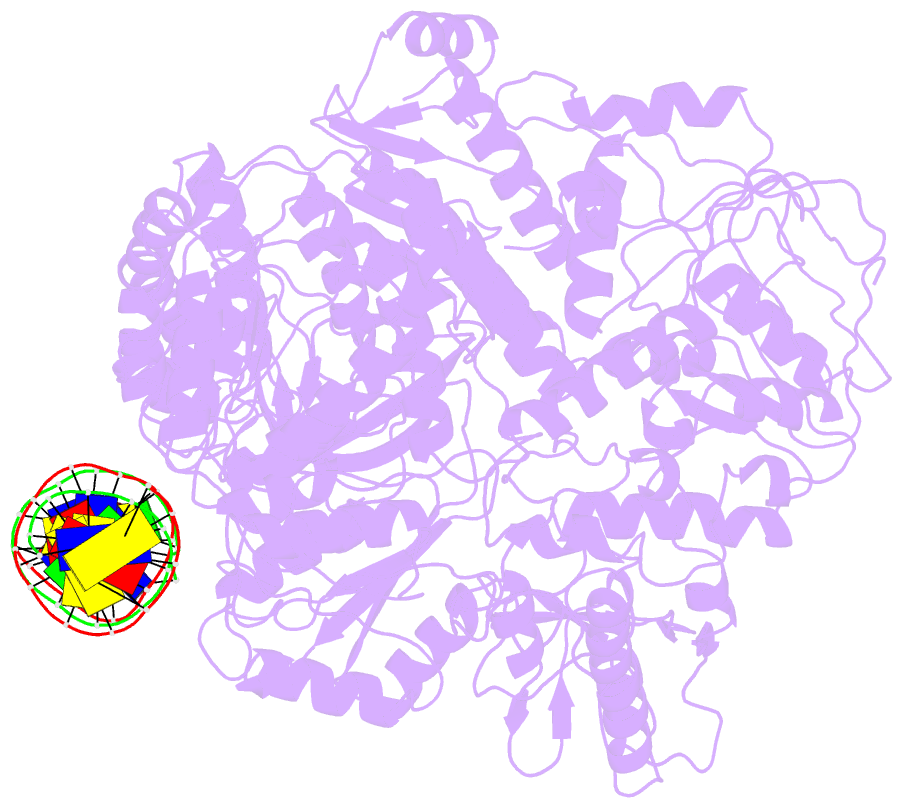
- Structure of mfd bound to dsDNA
- Reference
-
Brugger C, Zhang C, Suhanovsky MM, Kim DD, Sinclair AN,
Lyumkis D, Deaconescu AM (2020): "Molecular
determinants for dsDNA translocation by the
transcription-repair coupling and evolvability factor
Mfd." Nat Commun, 11, 3740.
doi: 10.1038/s41467-020-17457-1.
- Abstract
- Mfd couples transcription to nucleotide excision
repair, and acts on RNA polymerases when elongation is
impeded. Depending on impediment severity, this action
results in either transcription termination or elongation
rescue, which rely on ATP-dependent Mfd translocation on
DNA. Due to its role in antibiotic resistance, Mfd is also
emerging as a prime target for developing anti-evolution
drugs. Here we report the structure of DNA-bound Mfd, which
reveals large DNA-induced structural changes that are
linked to the active site via ATPase motif VI. These
changes relieve autoinhibitory contacts between the N- and
C-termini and unmask UvrA recognition determinants. We also
demonstrate that translocation relies on a threonine in
motif Ic, widely conserved in translocases, and a
family-specific histidine near motif IVa, reminiscent of
the "arginine clamp" of RNA helicases. Thus, Mfd employs a
mode of DNA recognition that at its core is common to ss/ds
translocases that act on DNA or RNA.