Summary information and primary citation
- PDB-id
- 6xez; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-hydrolase-RNA
- Method
- cryo-EM (3.5 Å)
- Summary
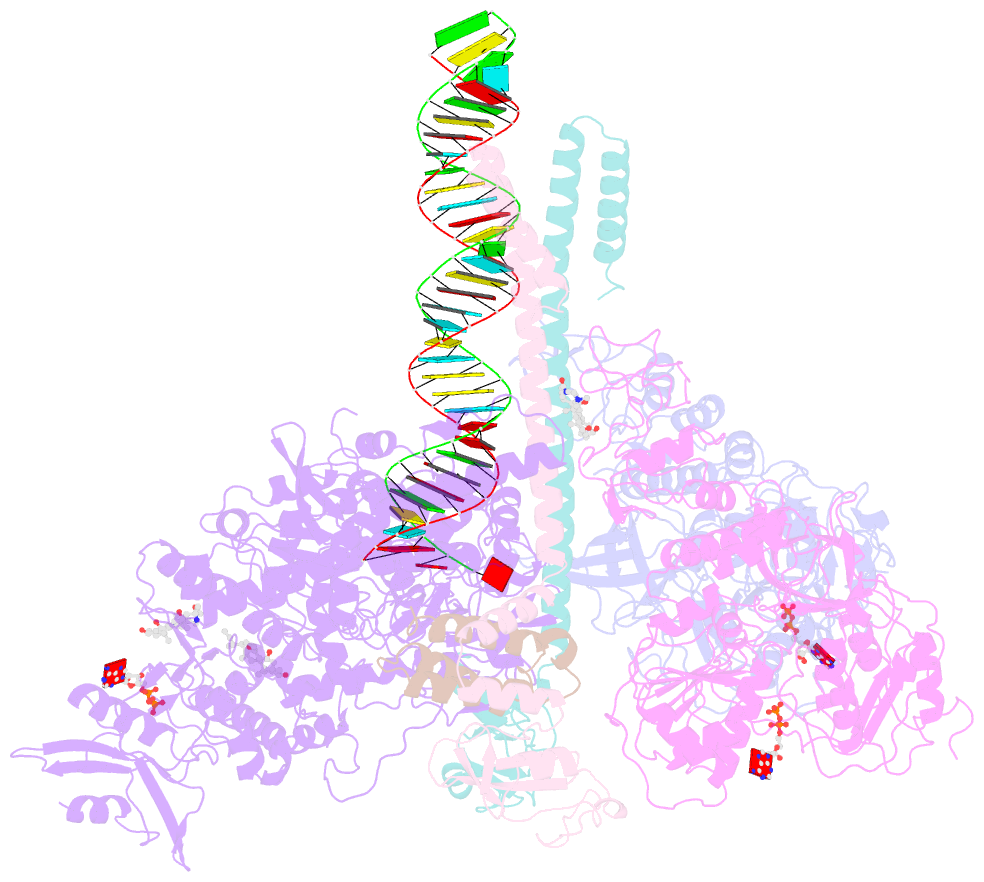
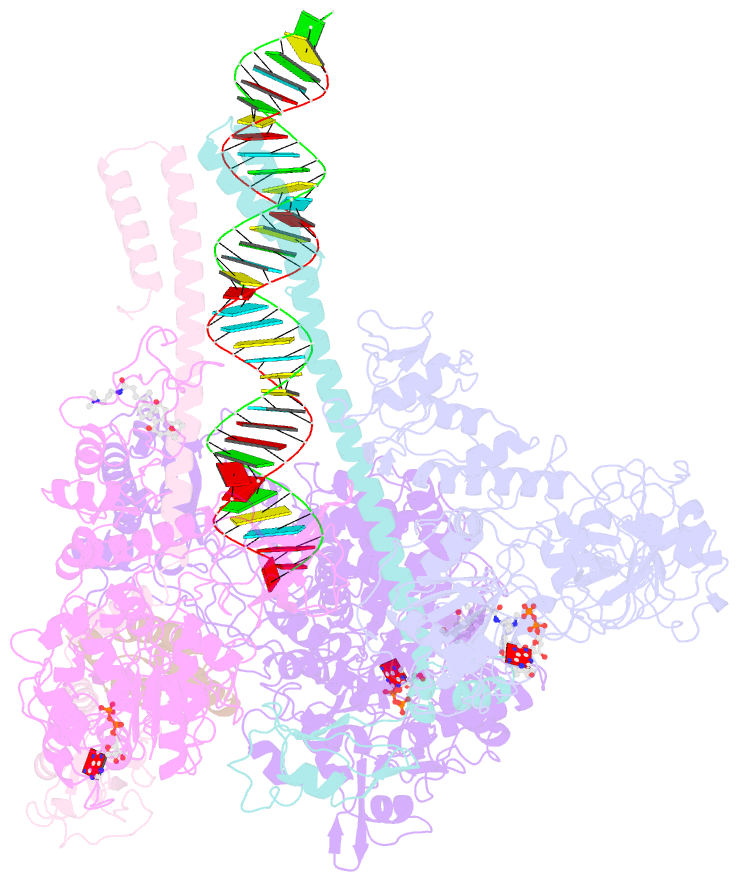
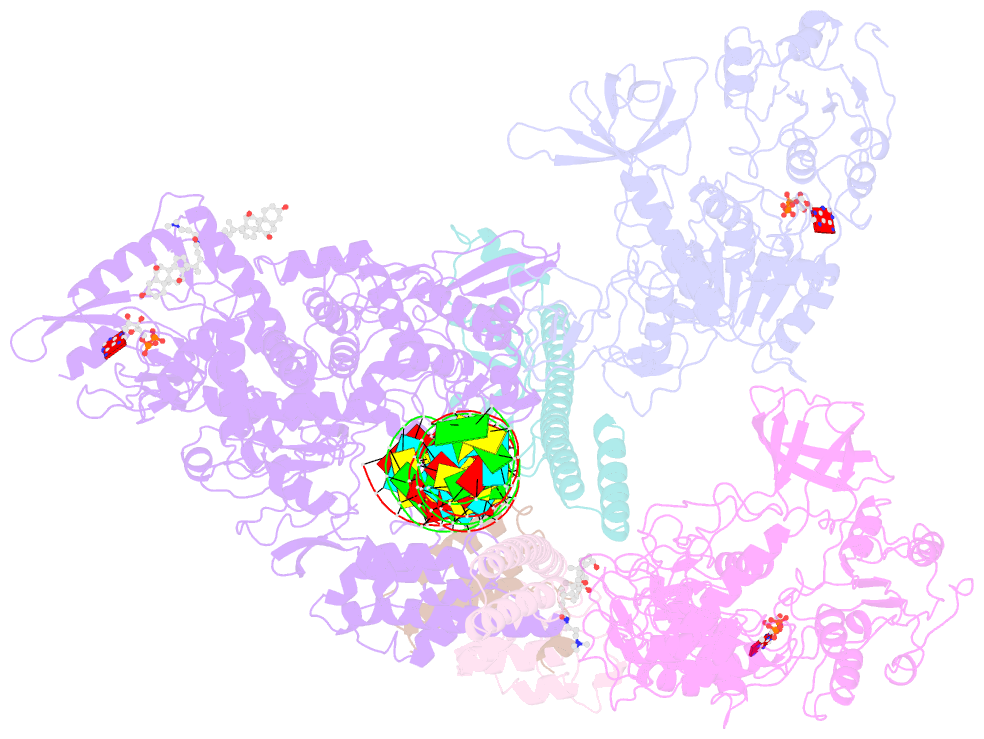
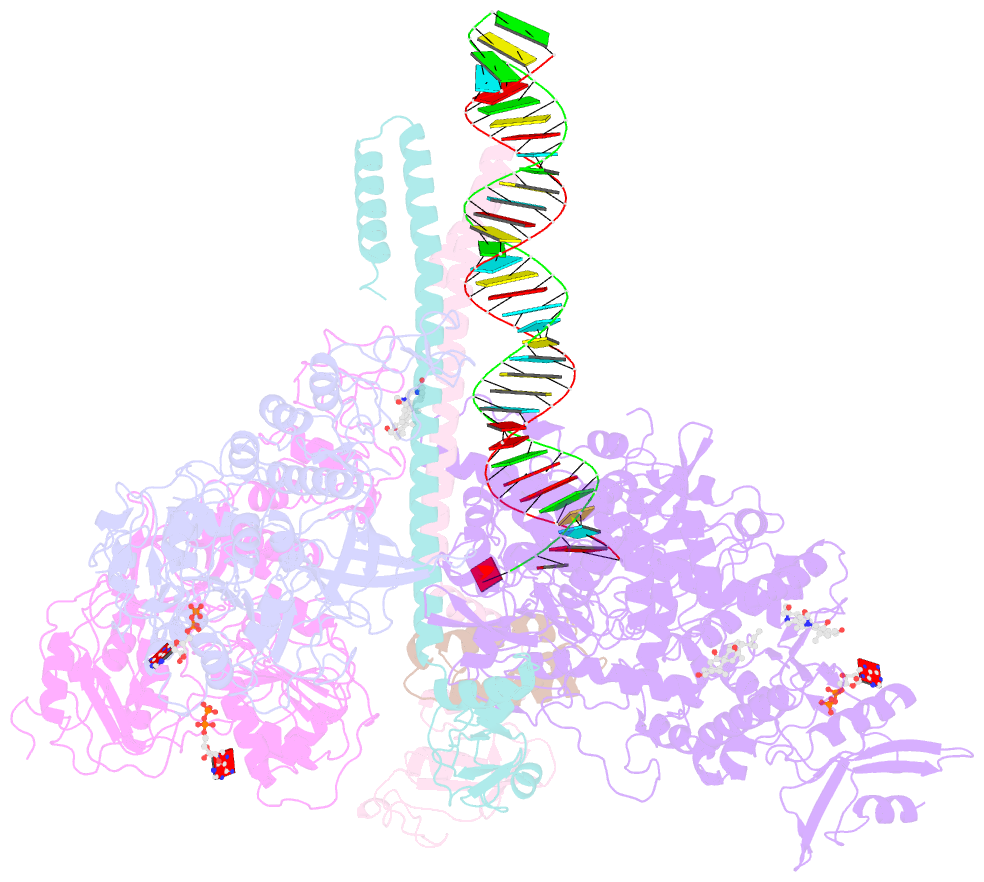
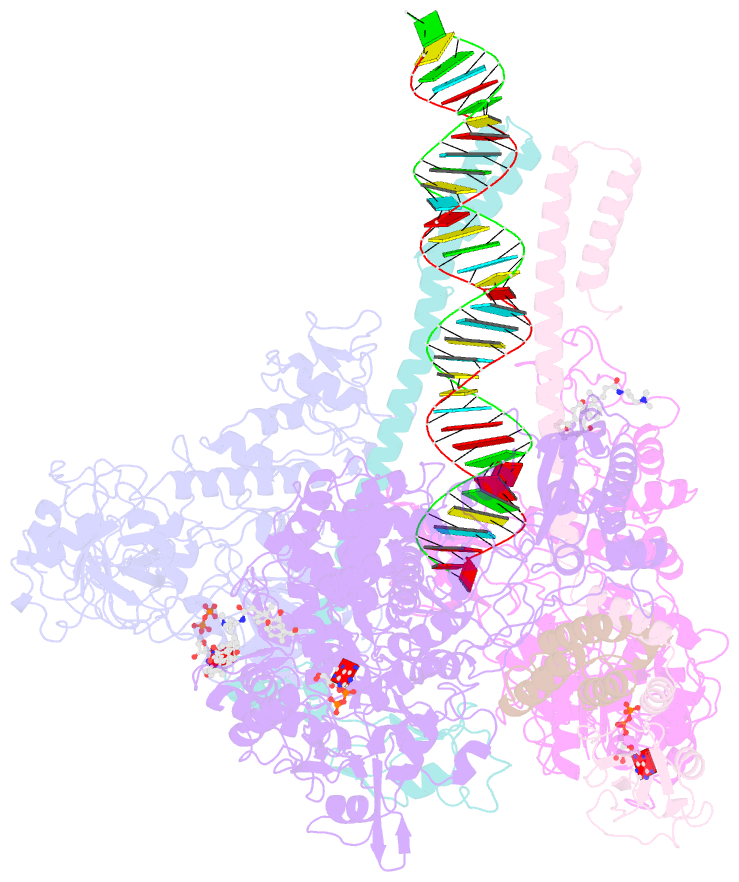
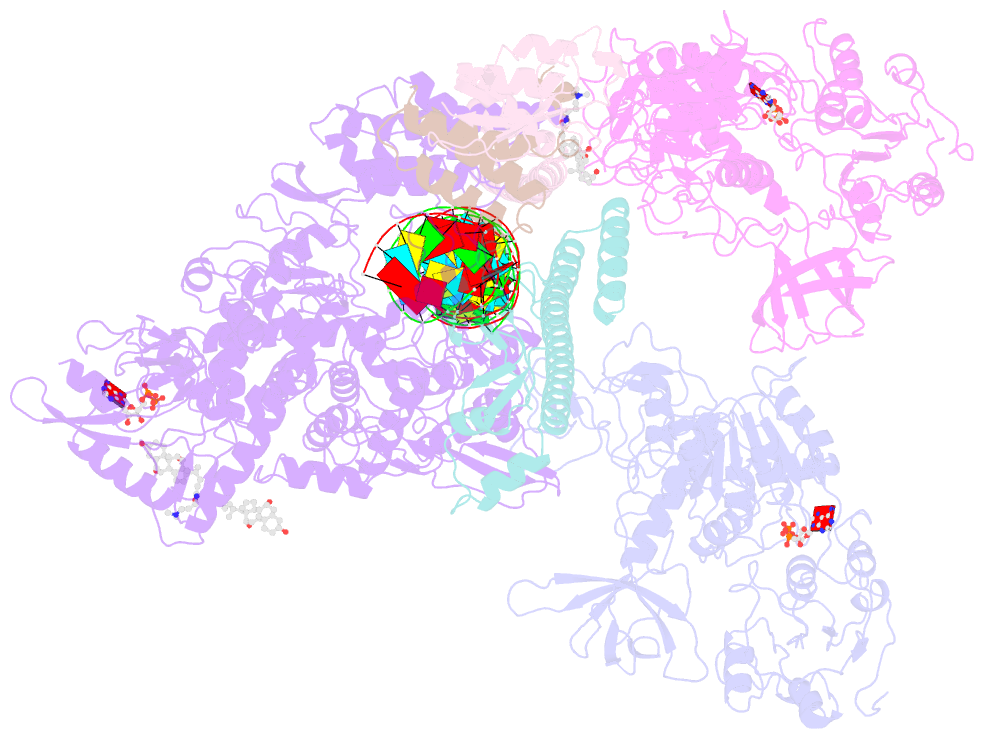
- Structure of sars-cov-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-rtc
- Reference
- Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, Maruthi K, Eng ET, Vatandaslar H, Chait BT, Kapoor TM, Darst SA, Campbell EA (2020): "Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex." Cell, 182, 1560-1573.e13. doi: 10.1016/j.cell.2020.07.033.
- Abstract
- SARS-CoV-2 is the causative agent of the 2019-2020 pandemic. The SARS-CoV-2 genome is replicated and transcribed by the RNA-dependent RNA polymerase holoenzyme (subunits nsp7/nsp82/nsp12) along with a cast of accessory factors. One of these factors is the nsp13 helicase. Both the holo-RdRp and nsp13 are essential for viral replication and are targets for treating the disease COVID-19. Here we present cryoelectron microscopic structures of the SARS-CoV-2 holo-RdRp with an RNA template product in complex with two molecules of the nsp13 helicase. The Nidovirales order-specific N-terminal domains of each nsp13 interact with the N-terminal extension of each copy of nsp8. One nsp13 also contacts the nsp12 thumb. The structure places the nucleic acid-binding ATPase domains of the helicase directly in front of the replicating-transcribing holo-RdRp, constraining models for nsp13 function. We also observe ADP-Mg2+ bound in the nsp12 N-terminal nidovirus RdRp-associated nucleotidyltransferase domain, detailing a new pocket for anti-viral therapy development.