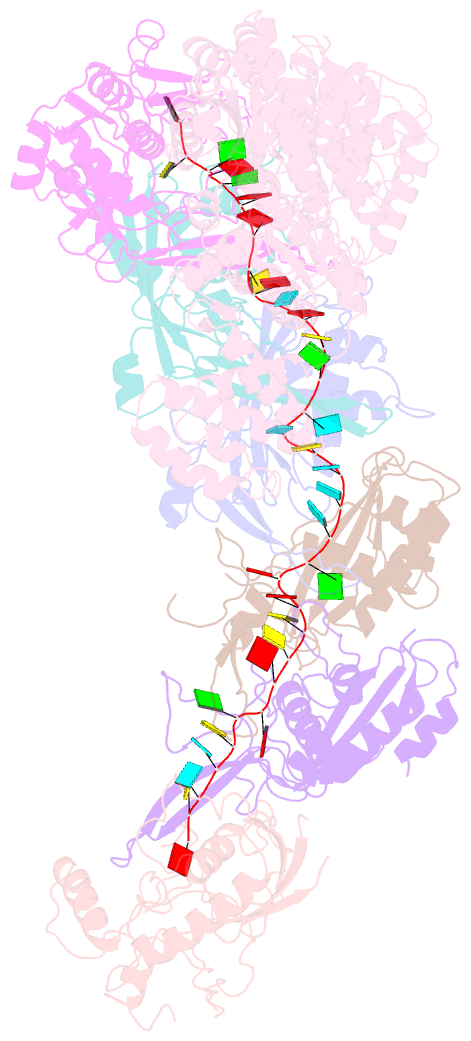
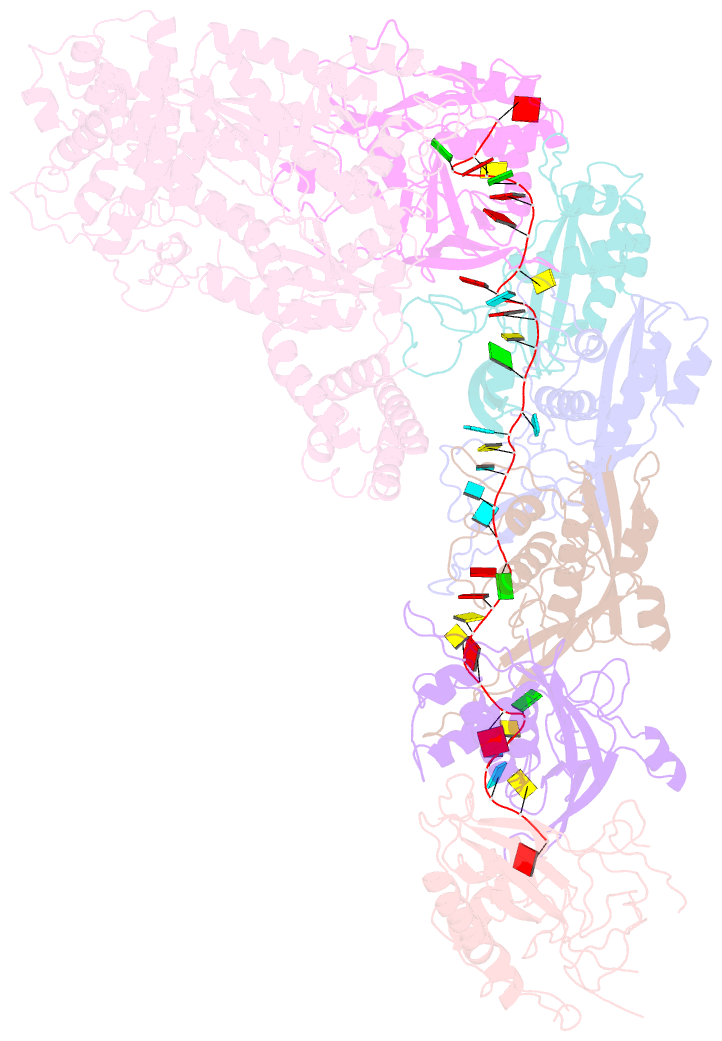
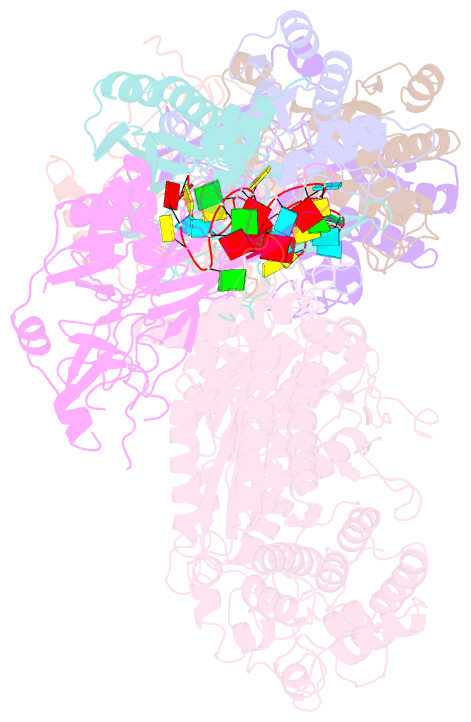
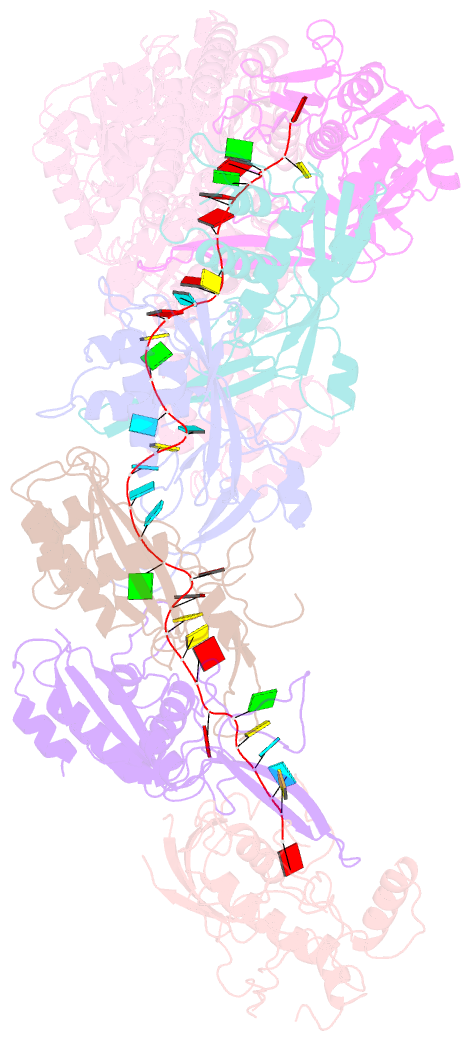
Summary information and primary citation
- PDB-id
- 6xn5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (2.97 Å)
- Summary
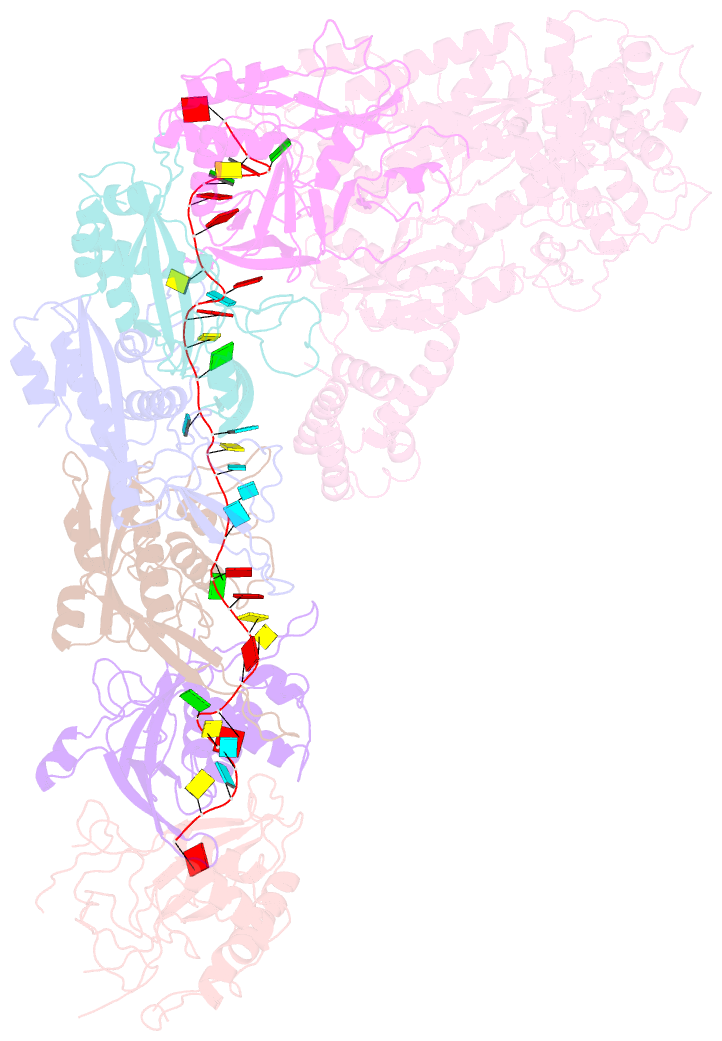
- Structure of the lactococcus lactis csm apo- crispr-cas complex
- Reference
- Sridhara S, Rai J, Whyms C, Goswami H, He H, Woodside W, Terns MP, Li H (2022): "Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex." Commun Biol, 5, 279. doi: 10.1038/s42003-022-03187-1.
- Abstract
- The small RNA-mediated immunity in bacteria depends on foreign RNA-activated and self RNA-inhibited enzymatic activities. The multi-subunit Type III-A CRISPR-Cas effector complex (Csm) exemplifies this principle and is in addition regulated by cellular metabolites such as divalent metals and ATP. Recognition of the foreign or cognate target RNA (CTR) triggers its single-stranded deoxyribonuclease (DNase) and cyclic oligoadenylate (cOA) synthesis activities. The same activities remain dormant in the presence of the self or non-cognate target RNA (NTR) that differs from CTR only in its 3'-protospacer flanking sequence (3'-PFS). Here we employ electron cryomicroscopy (cryoEM), functional assays, and comparative cross-linking to study in vivo assembled mesophilic Lactococcus lactis Csm (LlCsm) at the three functional states: apo, the CTR- and the NTR-bound. Unlike previously studied Csm complexes, we observed binding of 3'-PFS to Csm in absence of bound ATP and analyzed the structures of the four RNA cleavage sites. Interestingly, comparative crosslinking results indicate a tightening of the Csm3-Csm4 interface as a result of CTR but not NTR binding, reflecting a possible role of protein dynamics change during activation.