Summary information and primary citation
- PDB-id
- 6y4b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (5.0 Å)
- Summary
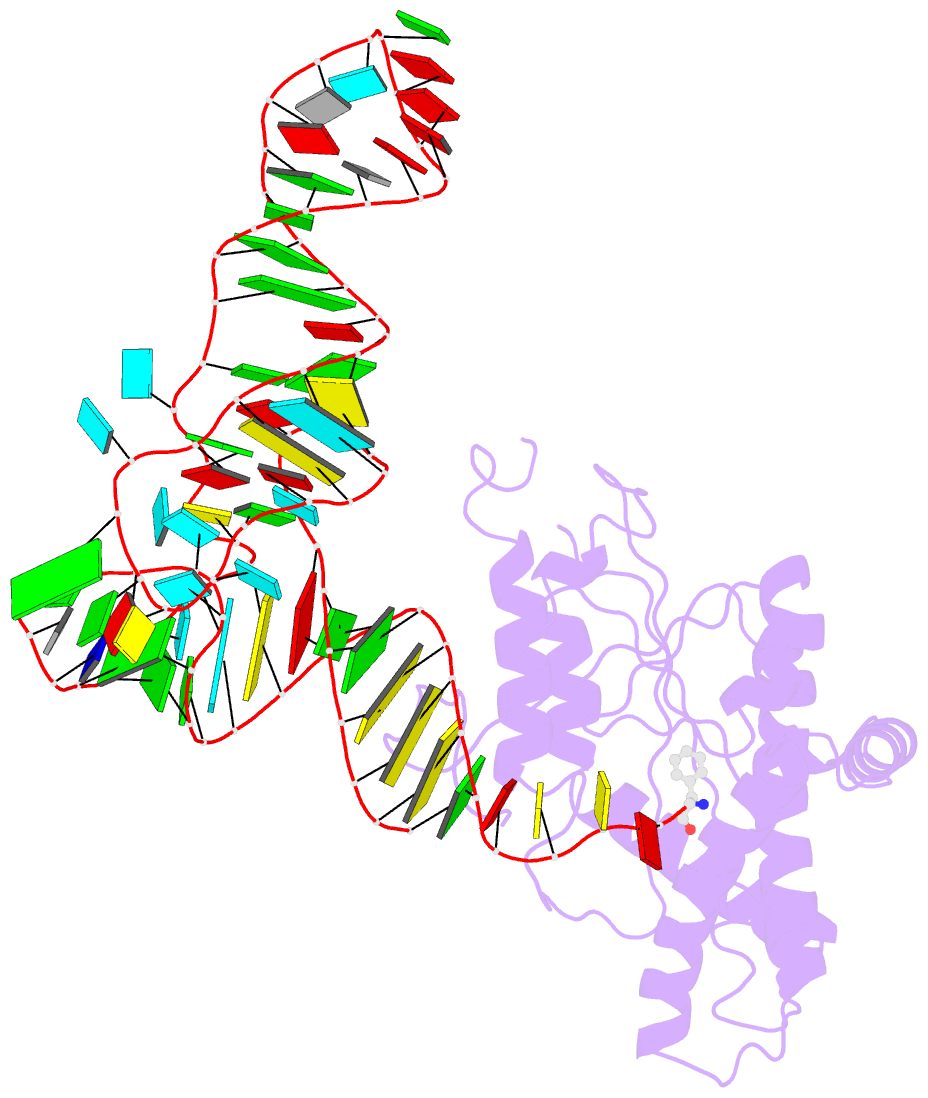
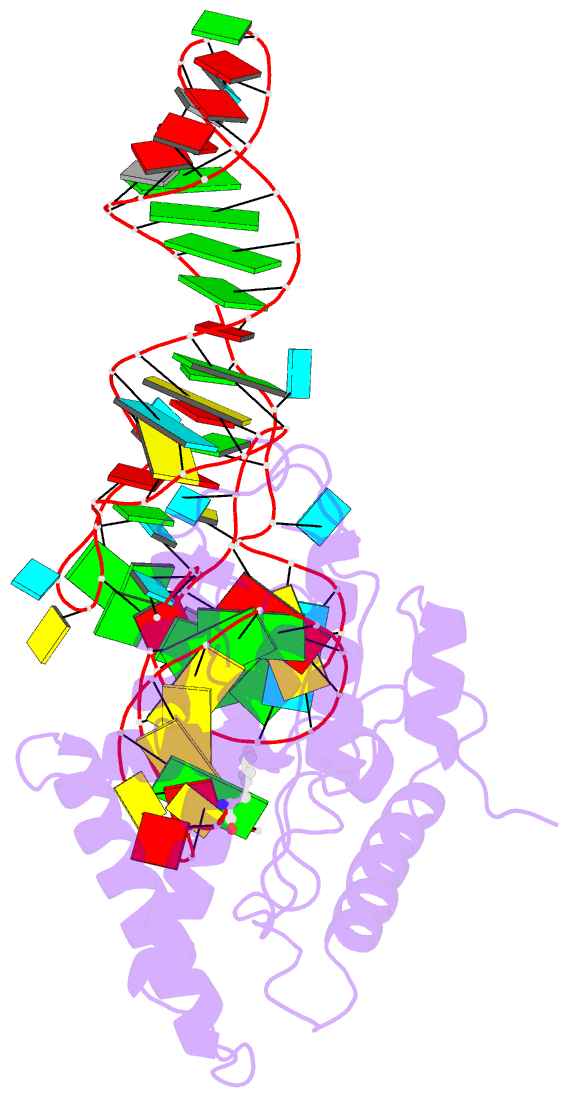
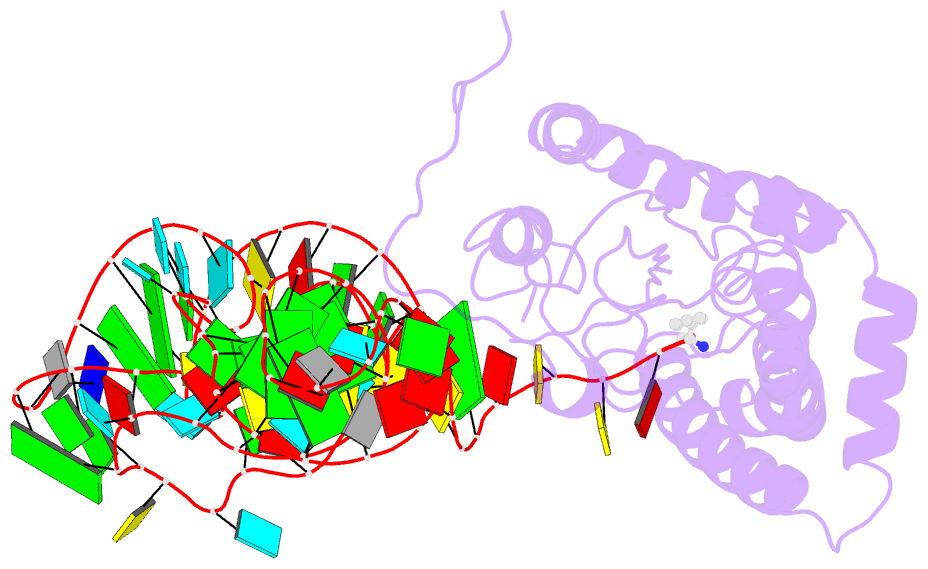
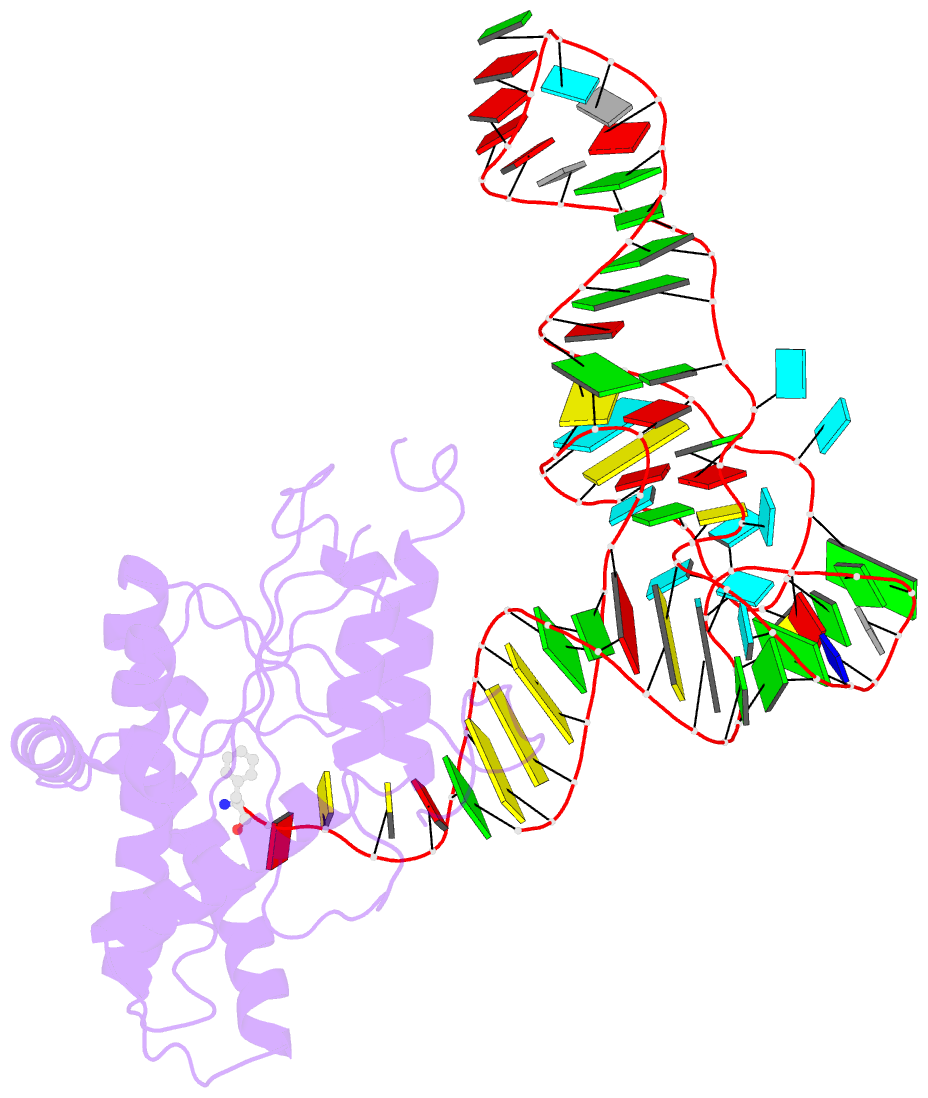
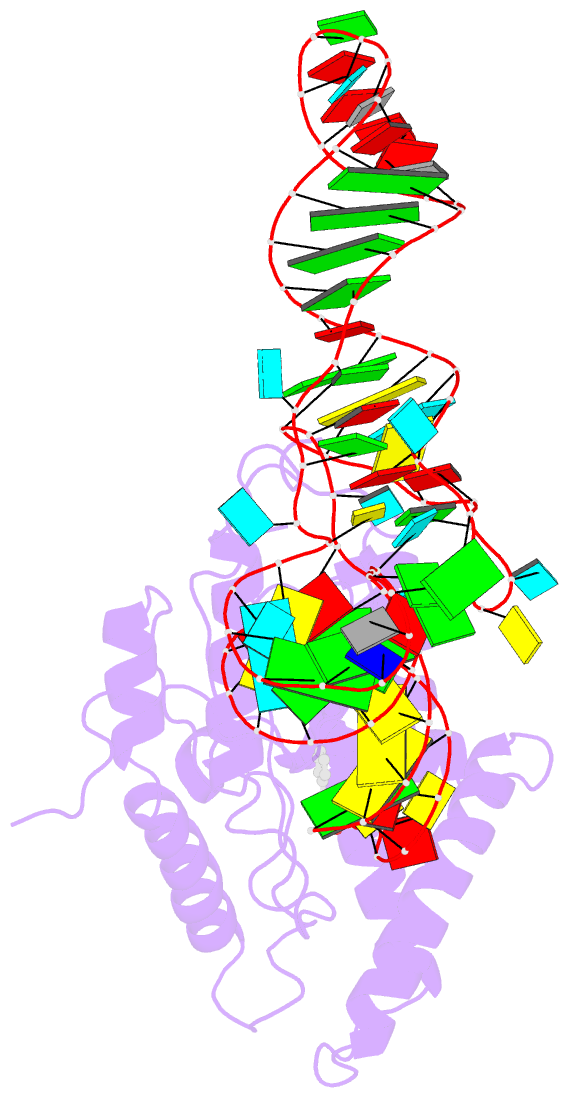
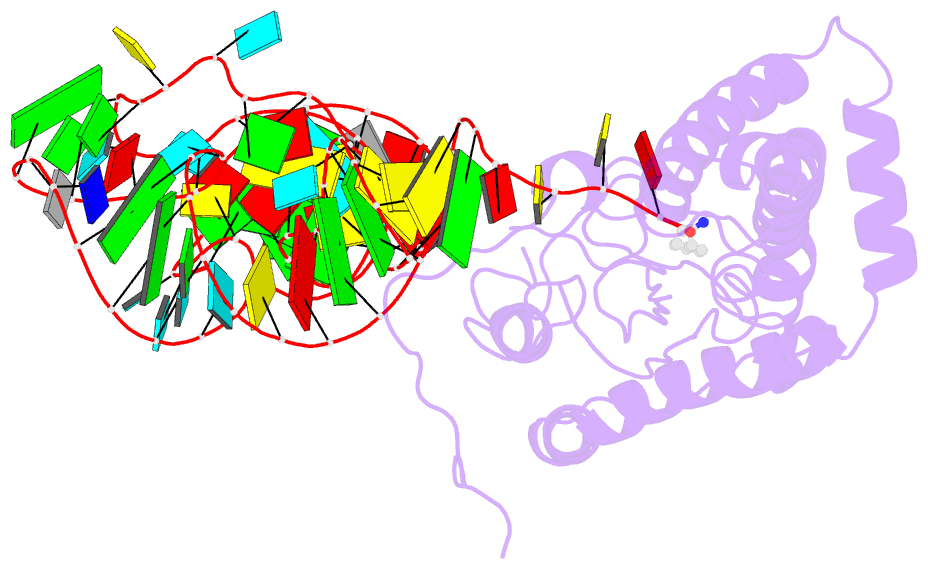
- Structure of cyclodipeptide synthase from candidatus glomeribacter gigasporarum bound to phe-trnaphe
- Reference
- Bourgeois G, Seguin J, Babin M, Gondry M, Mechulam Y, Schmitt E (2020): "Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates." Rna, 26, 1589-1602. doi: 10.1261/rna.075184.120.
- Abstract
- Cyclodipeptide synthases (CDPSs) catalyze the synthesis of various cyclodipeptides by using two aminoacyl-tRNA (aa-tRNA) substrates in a sequential mechanism. Here, we studied binding of phenylalanyl-tRNAPhe to the CDPS from Candidatus Glomeribacter gigasporarum (Cglo-CDPS) by gel filtration and electrophoretic mobility shift assay. We determined the crystal structure of the Cglo-CDPS:Phe-tRNAPhe complex to 5 Å resolution and further studied it in solution using small-angle X-ray scattering (SAXS). The data show that the major groove of the acceptor stem of the aa-tRNA interacts with the enzyme through the basic β2 and β7 strands of CDPSs belonging to the XYP subfamily. A bending of the CCA extremity enables the amino acid moiety to be positioned in the P1 pocket while the terminal A76 adenosine occupies the P2 pocket. Such a positioning indicates that the present structure illustrates the binding of the first aa-tRNA. In cells, CDPSs and the elongation factor EF-Tu share aminoacylated tRNAs as substrates. The present study shows that CDPSs and EF-Tu interact with opposite sides of tRNA. This may explain how CDPSs hijack aa-tRNAs from canonical ribosomal protein synthesis.