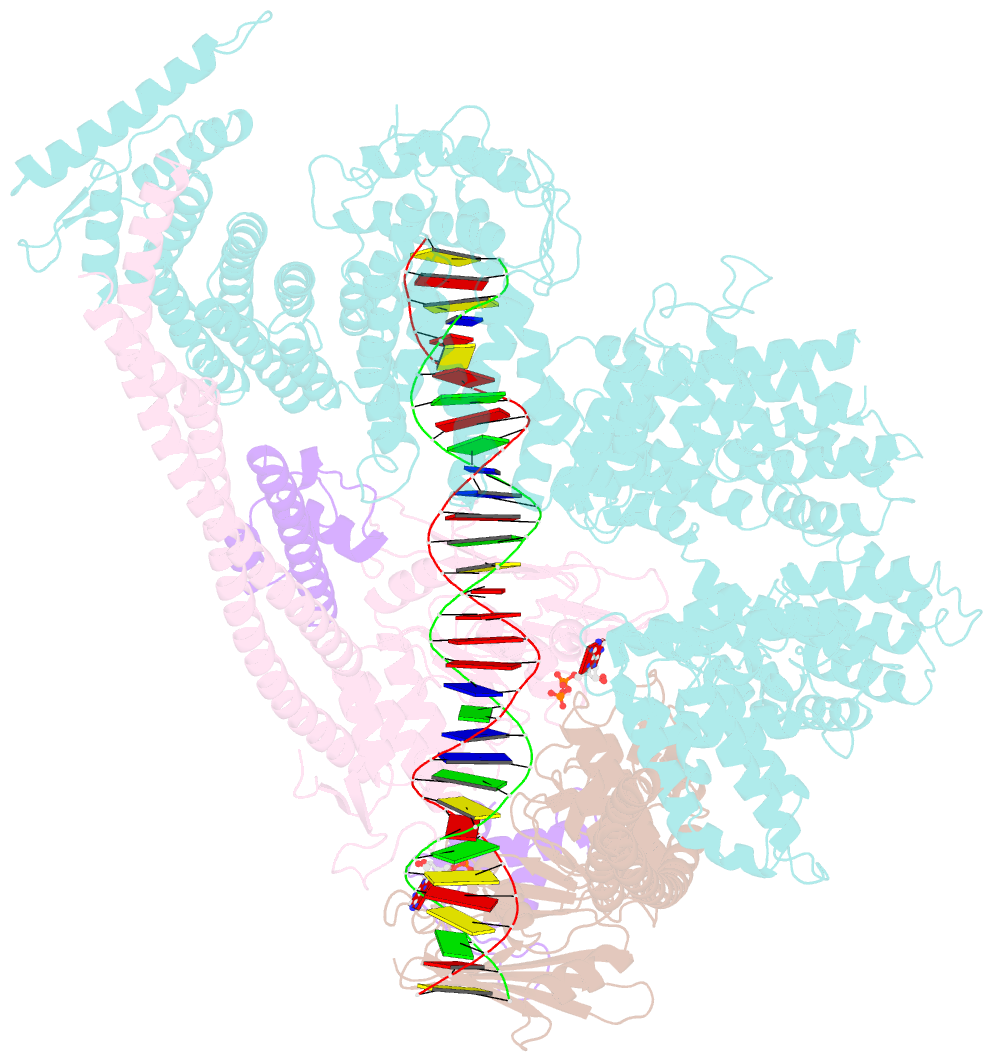
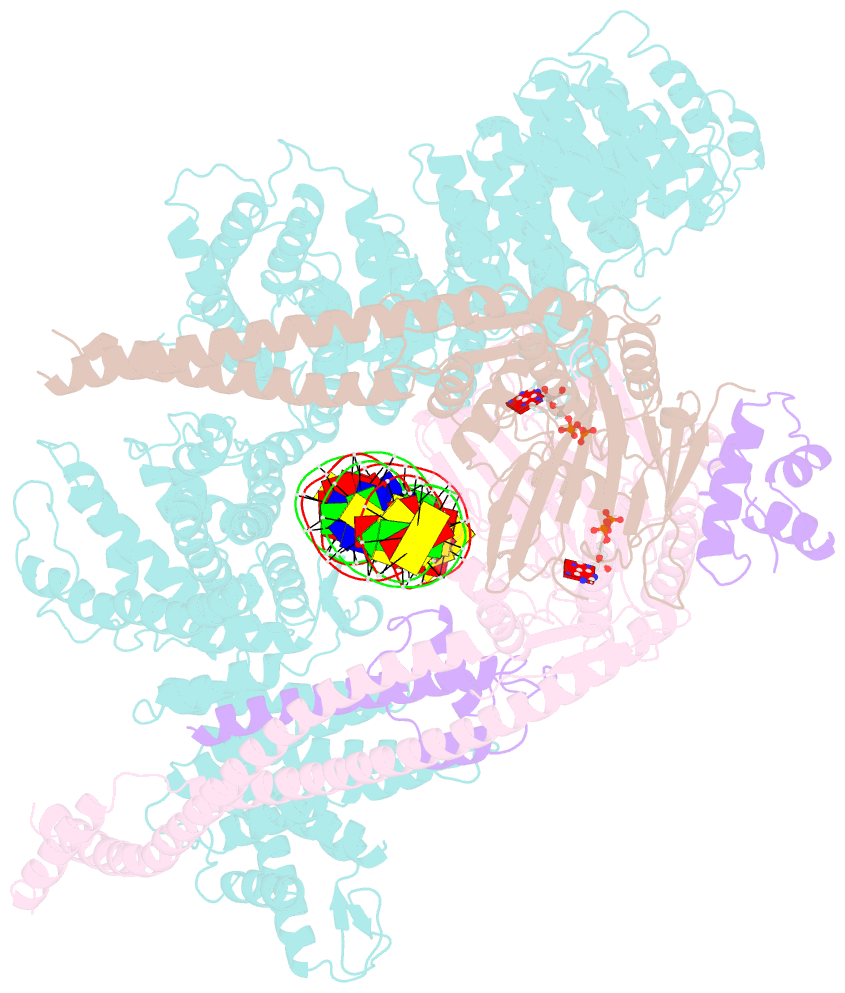
Summary information and primary citation
- PDB-id
- 6yuf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (3.94 Å)
- Summary
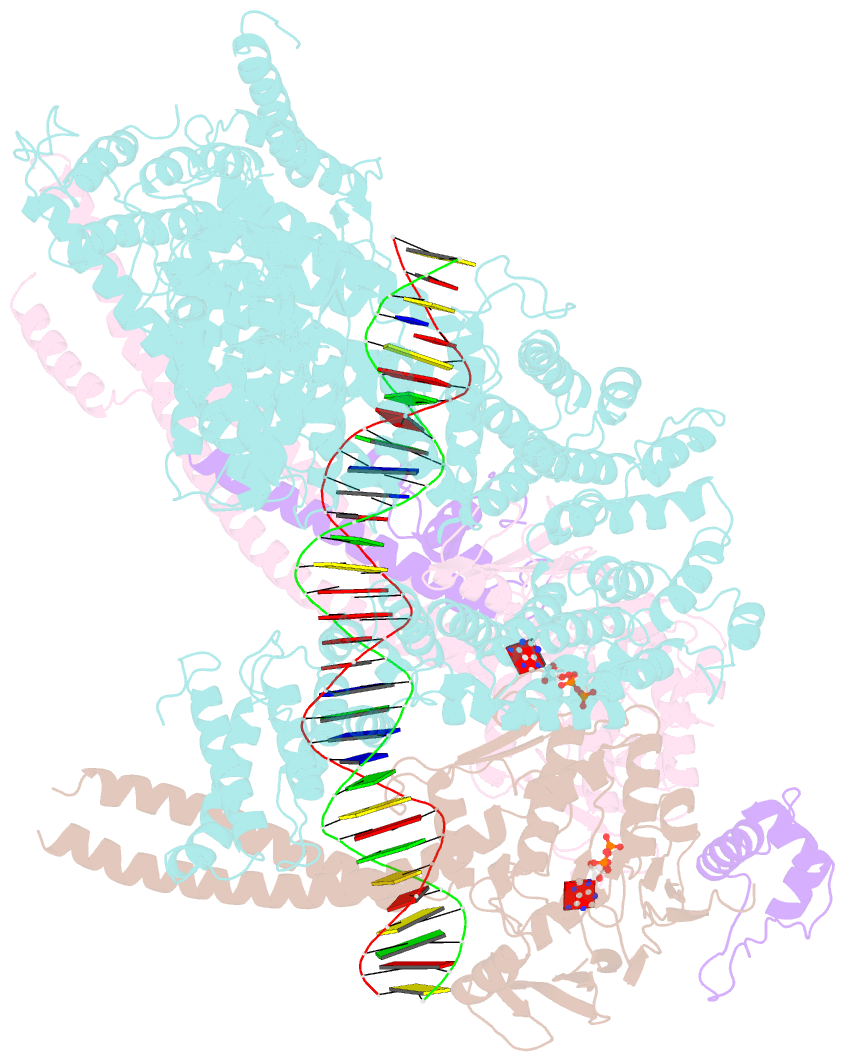
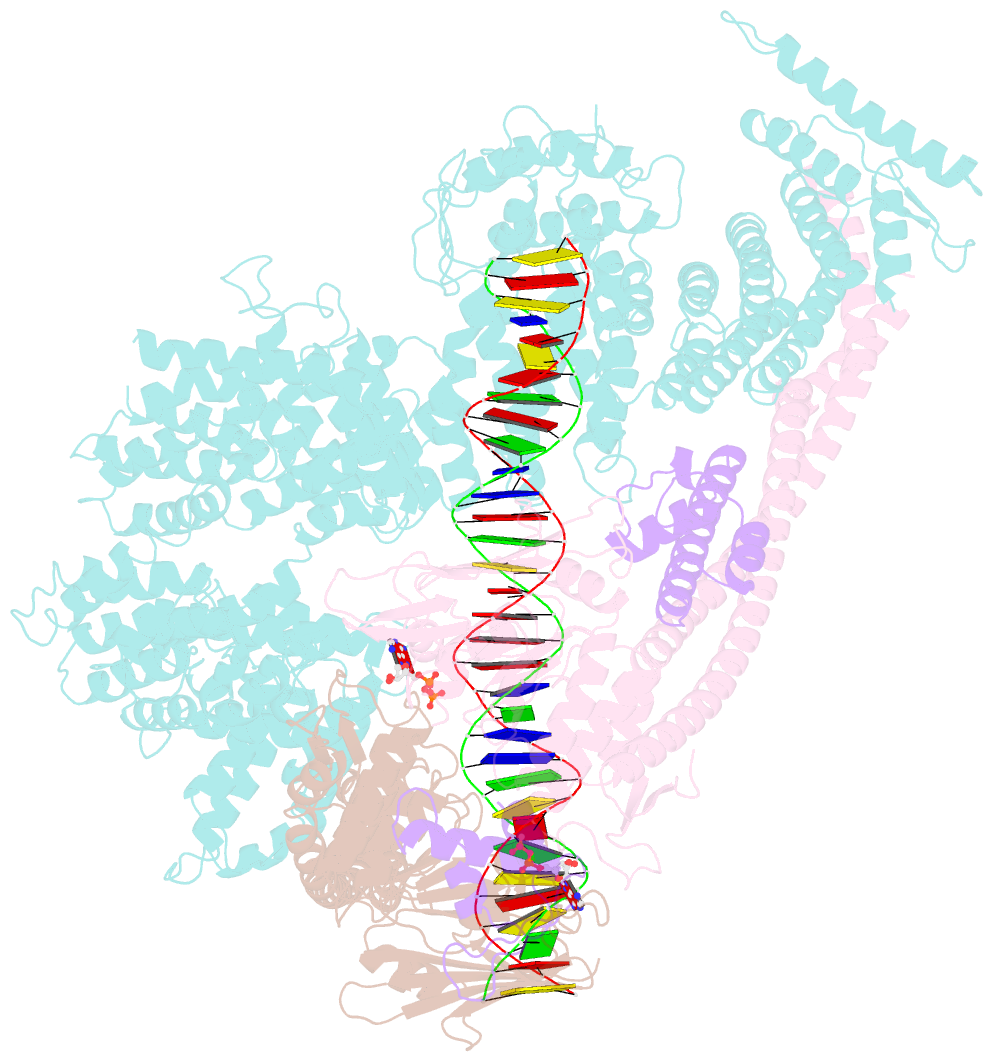
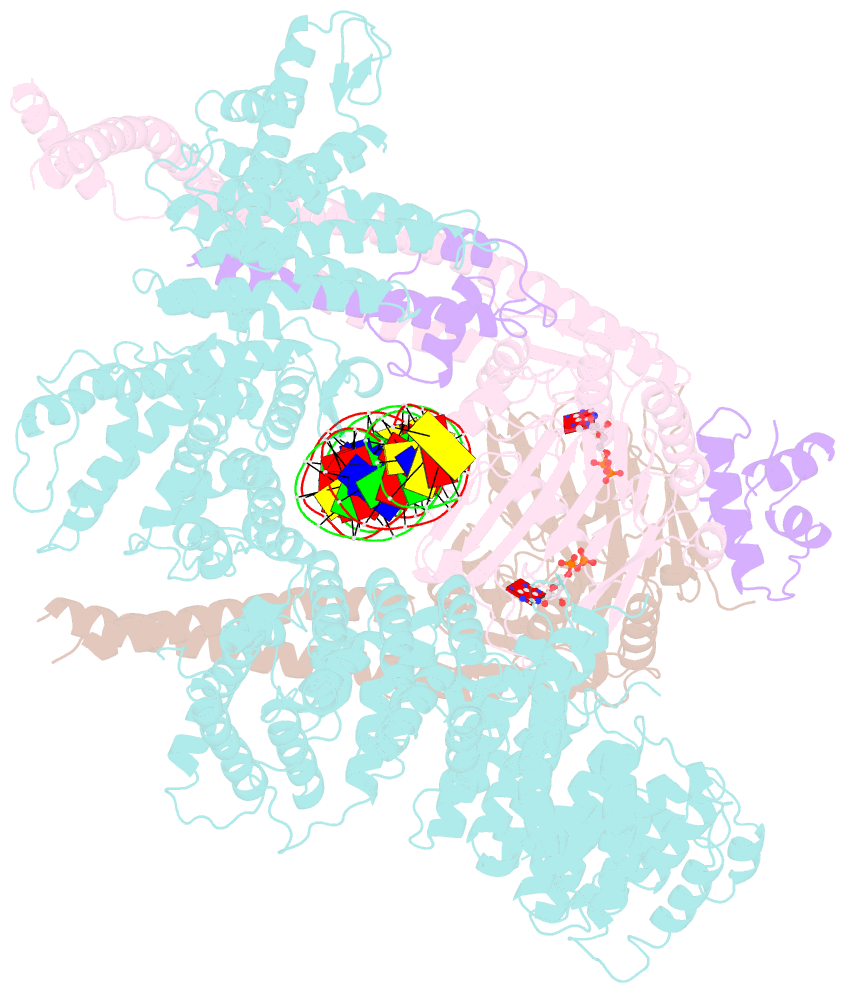
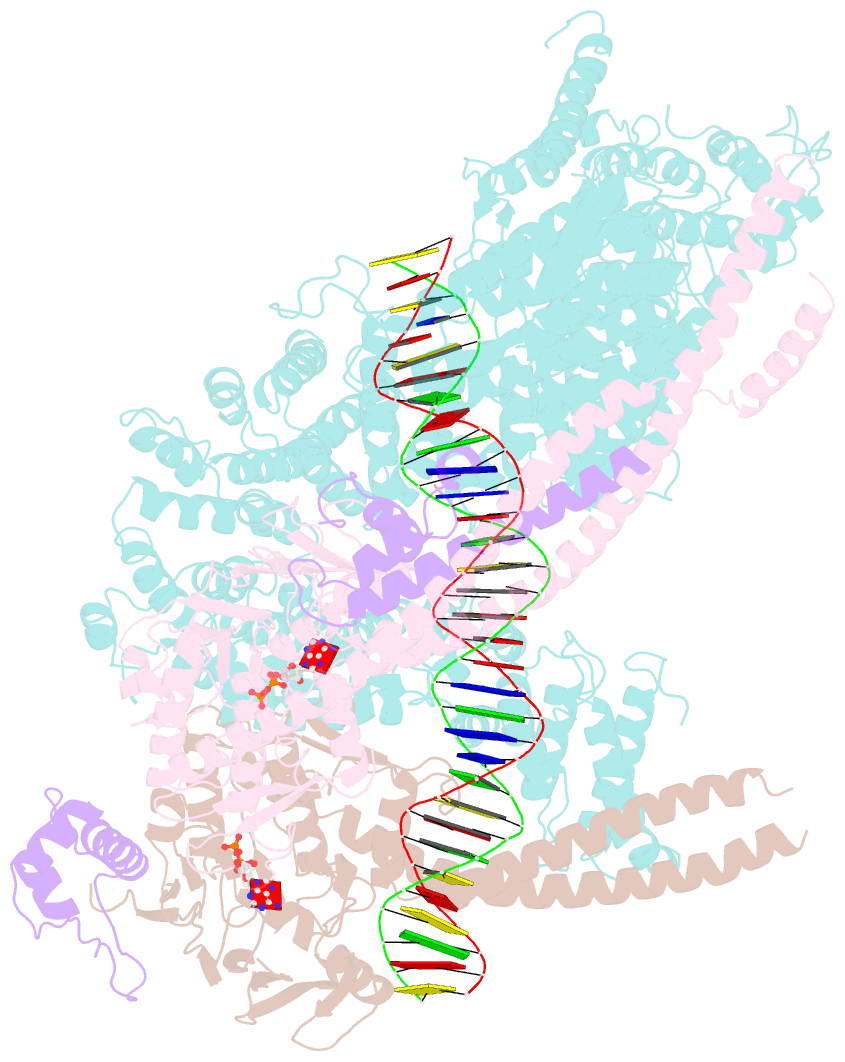
- Cohesin complex with loader gripping DNA
- Reference
- Higashi TL, Eickhoff P, Sousa JS, Locke J, Nans A, Flynn HR, Snijders AP, Papageorgiou G, O'Reilly N, Chen ZA, O'Reilly FJ, Rappsilber J, Costa A, Uhlmann F (2020): "A Structure-Based Mechanism for DNA Entry into the Cohesin Ring." Mol.Cell, 79, 917. doi: 10.1016/j.molcel.2020.07.013.
- Abstract
- Despite key roles in sister chromatid cohesion and chromosome organization, the mechanism by which cohesin rings are loaded onto DNA is still unknown. Here we combine biochemical approaches and cryoelectron microscopy (cryo-EM) to visualize a cohesin loading intermediate in which DNA is locked between two gates that lead into the cohesin ring. Building on this structural framework, we design experiments to establish the order of events during cohesin loading. In an initial step, DNA traverses an N-terminal kleisin gate that is first opened upon ATP binding and then closed as the cohesin loader locks the DNA against the ATPase gate. ATP hydrolysis will lead to ATPase gate opening to complete DNA entry. Whether DNA loading is successful or results in loop extrusion might be dictated by a conserved kleisin N-terminal tail that guides the DNA through the kleisin gate. Our results establish the molecular basis for cohesin loading onto DNA.