Summary information and primary citation
- PDB-id
- 6yyt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (2.9 Å)
- Summary
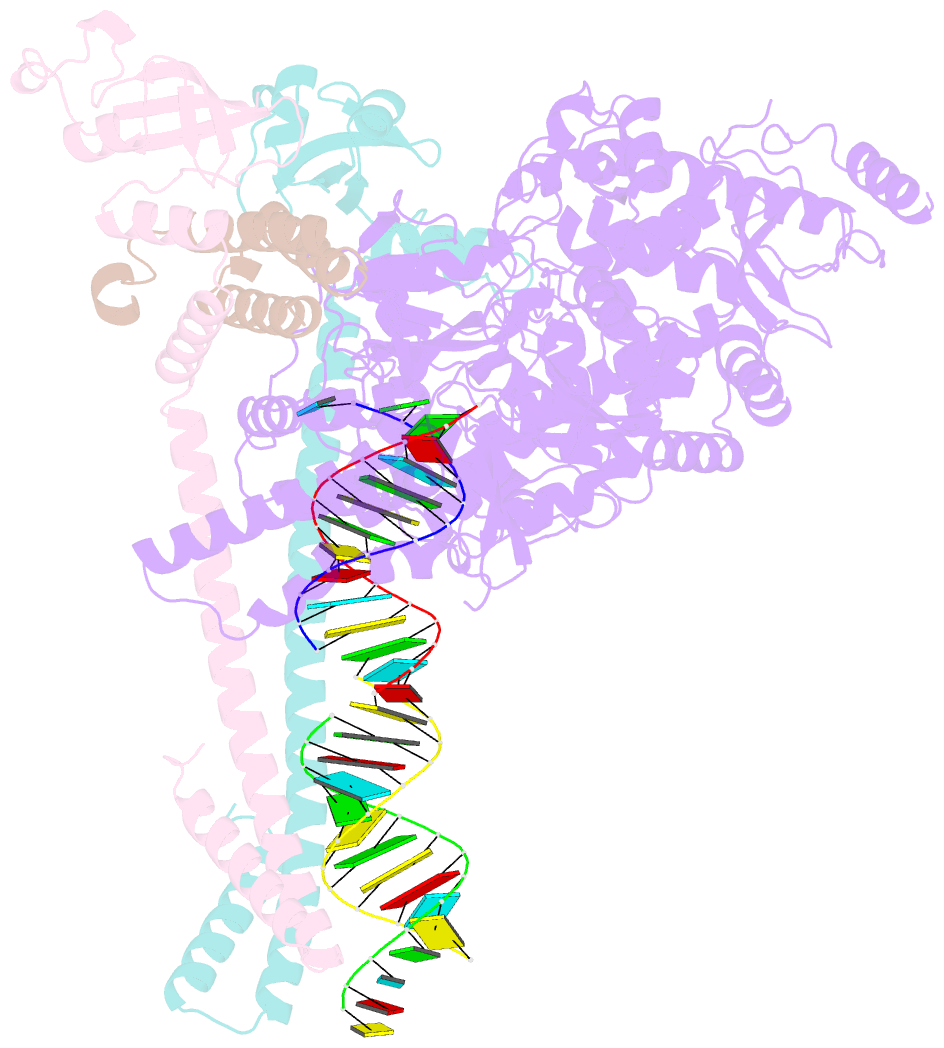
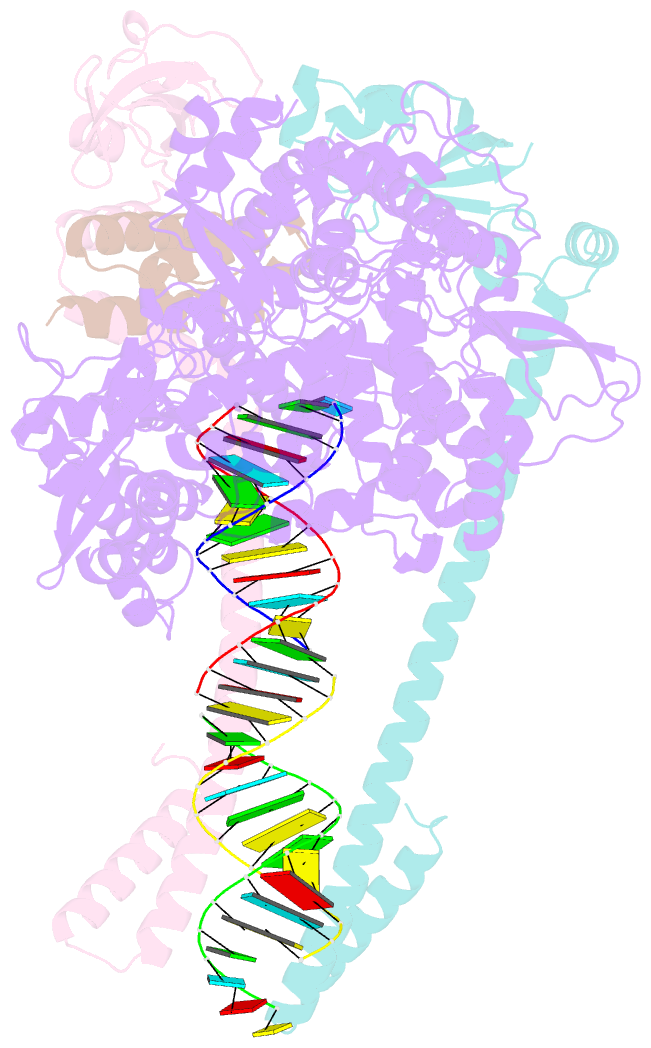
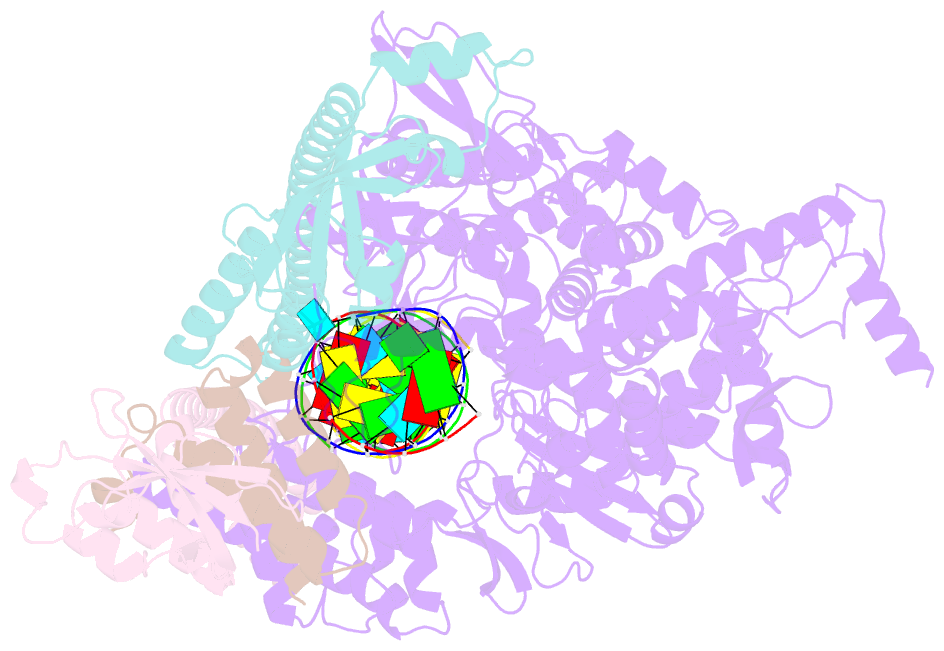
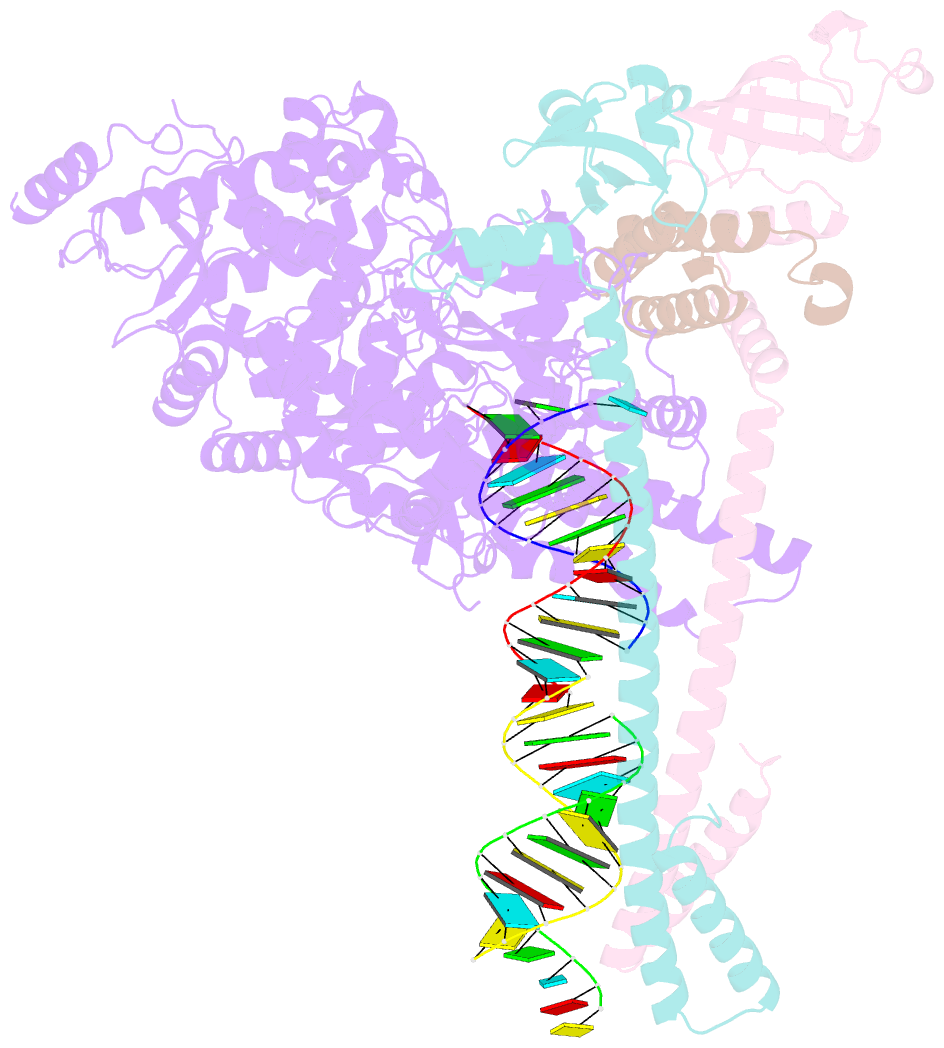
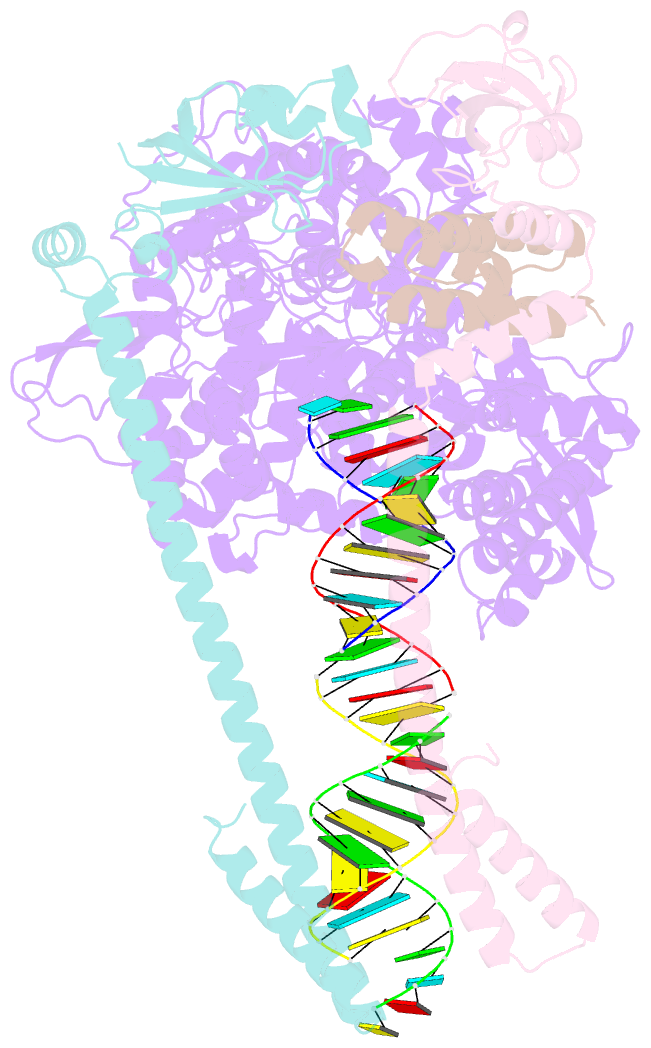
- Structure of replicating sars-cov-2 polymerase
- Reference
- Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P (2020): "Structure of replicating SARS-CoV-2 polymerase." Nature, 584, 154-156. doi: 10.1038/s41586-020-2368-8.
- Abstract
- The new coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) uses an RNA-dependent RNA polymerase (RdRp) for the replication of its genome and the transcription of its genes1-3. Here we present a cryo-electron microscopy structure of the SARS-CoV-2 RdRp in an active form that mimics the replicating enzyme. The structure comprises the viral proteins non-structural protein 12 (nsp12), nsp8 and nsp7, and more than two turns of RNA template-product duplex. The active-site cleft of nsp12 binds to the first turn of RNA and mediates RdRp activity with conserved residues. Two copies of nsp8 bind to opposite sides of the cleft and position the second turn of RNA. Long helical extensions in nsp8 protrude along exiting RNA, forming positively charged 'sliding poles'. These sliding poles can account for the known processivity of RdRp that is required for replicating the long genome of coronaviruses3. Our results enable a detailed analysis of the inhibitory mechanisms that underlie the antiviral activity of substances such as remdesivir, a drug for the treatment of coronavirus disease 2019 (COVID-19)4.