Summary information and primary citation
- PDB-id
- 6z1a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase
- Method
- X-ray (2.3 Å)
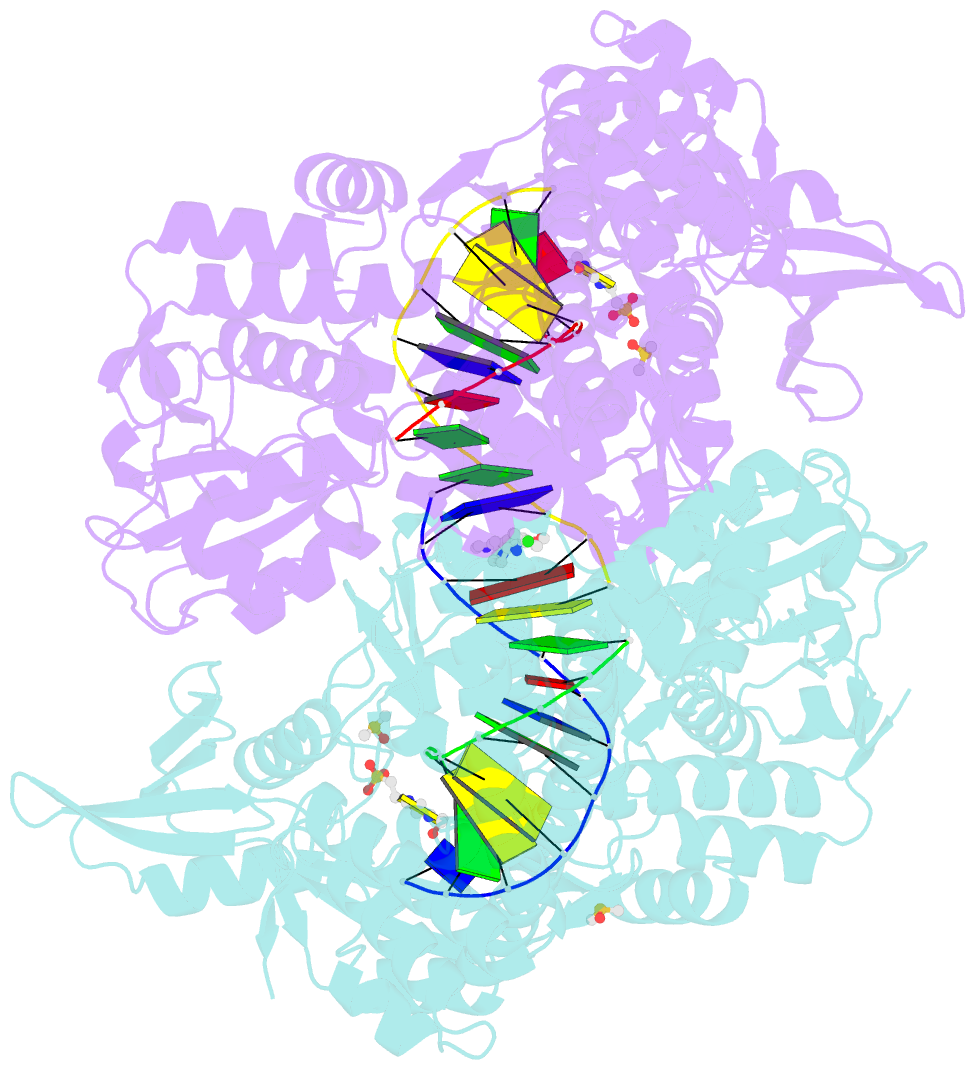
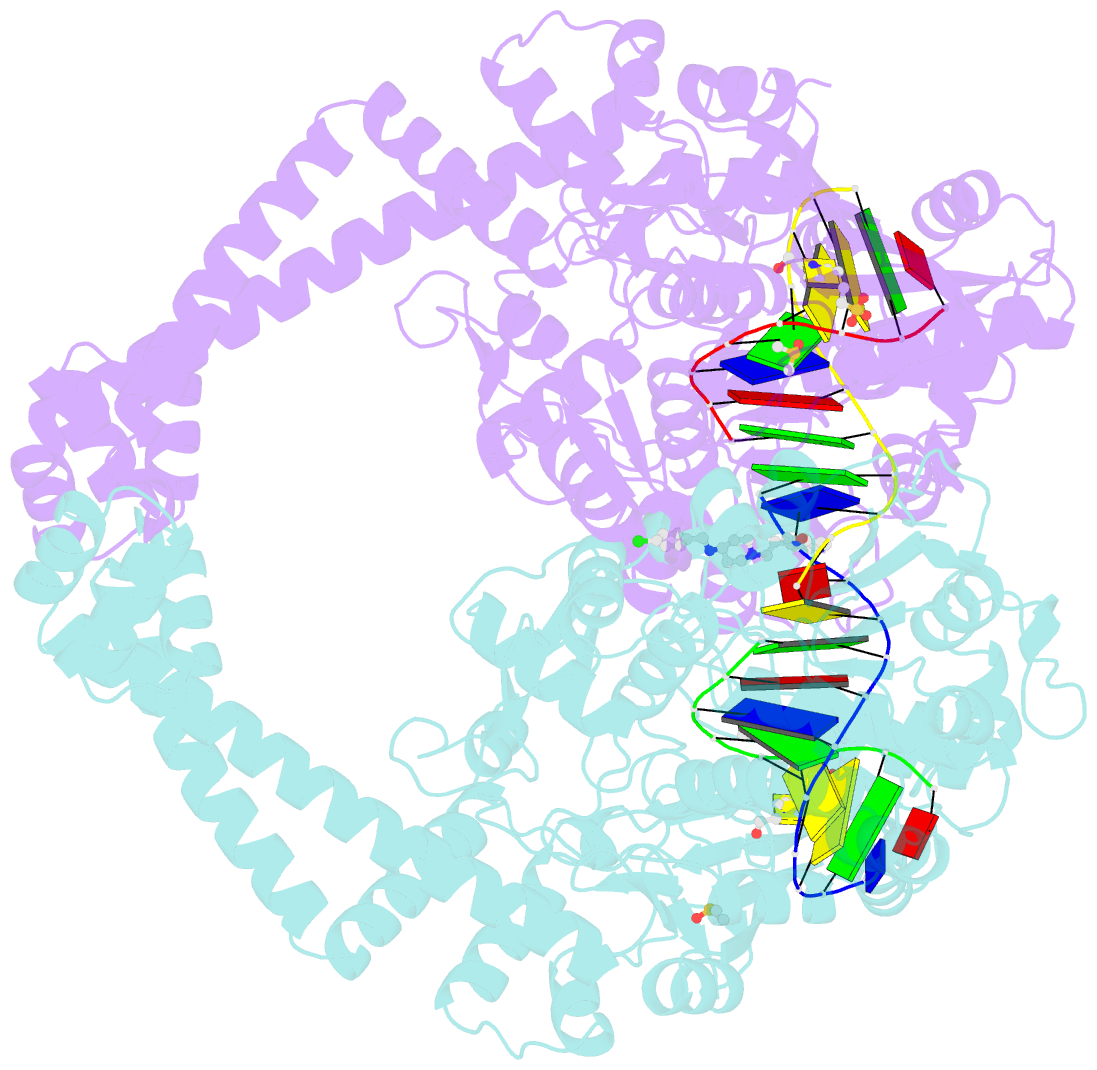
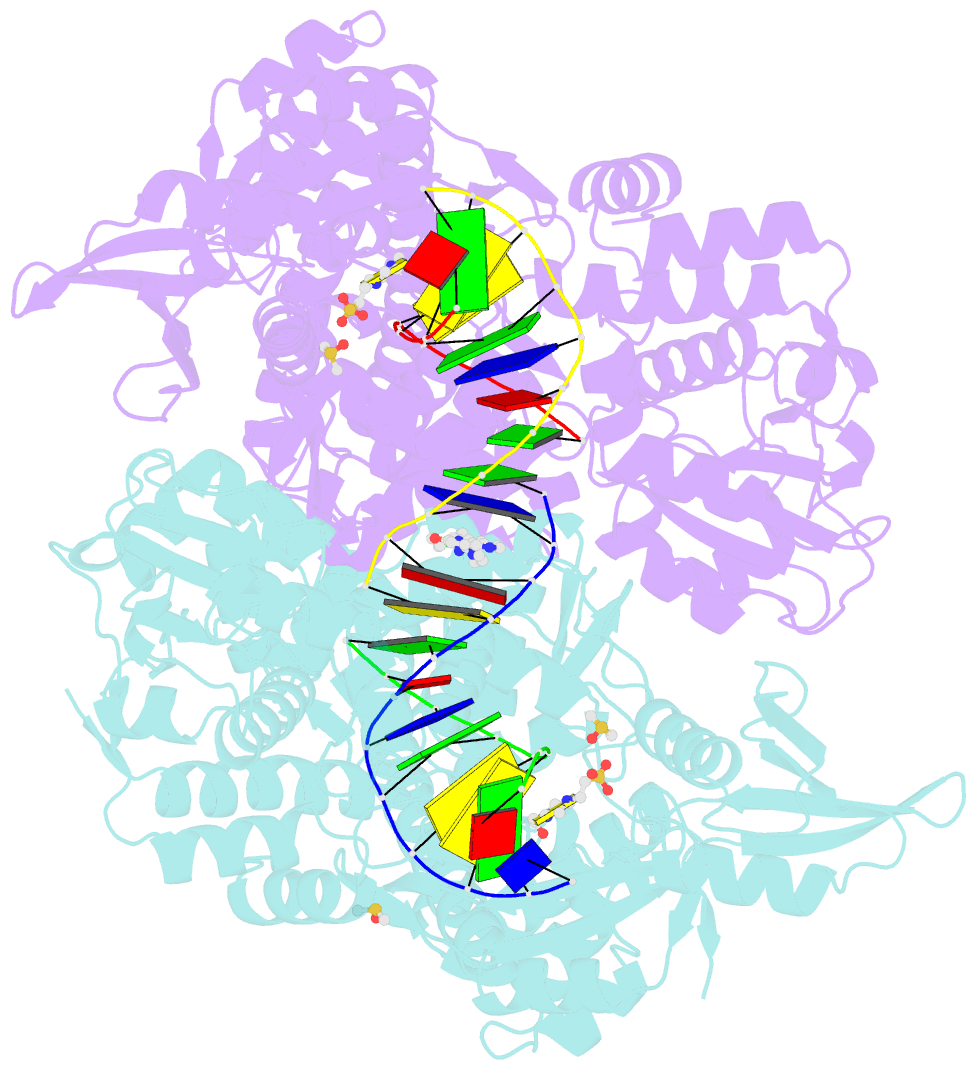
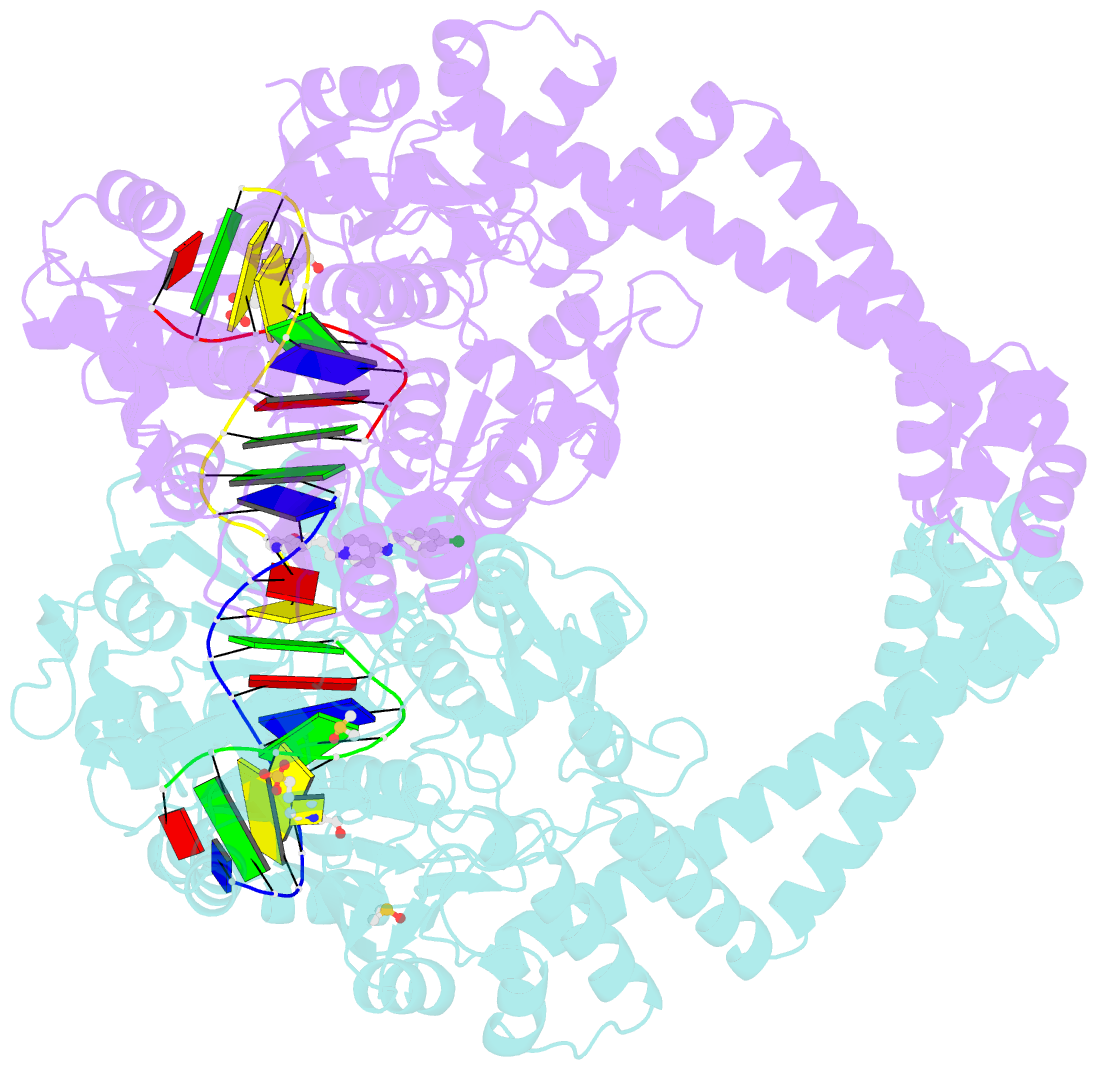
- Summary
- Ternary complex of staphylococcus aureus DNA gyrase with amk12 and DNA
- Reference
- Kolaric A, Germe T, Hrast M, Stevenson CEM, Lawson DM, Burton NP, Voros J, Maxwell A, Minovski N, Anderluh M (2021): "Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds." Nat Commun, 12, 150. doi: 10.1038/s41467-020-20405-8.
- Abstract
- Novel bacterial type II topoisomerase inhibitors (NBTIs) stabilize single-strand DNA cleavage breaks by DNA gyrase but their exact mechanism of action has remained hypothetical until now. We have designed a small library of NBTIs with an improved DNA gyrase-binding moiety resulting in low nanomolar inhibition and very potent antibacterial activity. They stabilize single-stranded cleavage complexes and, importantly, we have obtained the crystal structure where an NBTI binds gyrase-DNA in a single conformation lacking apparent static disorder. This directly proves the previously postulated NBTI mechanism of action and shows that they stabilize single-strand cleavage through asymmetric intercalation with a shift of the scissile phosphate. This crystal stucture shows that the chlorine forms a halogen bond with the backbone carbonyls of the two symmetry-related Ala68 residues. To the best of our knowledge, such a so-called symmetrical bifurcated halogen bond has not been identified in a biological system until now.